TLR-2 Recognizes Propionibacterium acnes CAMP Factor 1 from Highly Inflammatory Strains
- PMID: 27902761
- PMCID: PMC5130237
- DOI: 10.1371/journal.pone.0167237
TLR-2 Recognizes Propionibacterium acnes CAMP Factor 1 from Highly Inflammatory Strains
Abstract
Background: Propionibacterium acnes (P. acnes) is an anaerobic, Gram-positive bacteria encountered in inflammatory acne lesions, particularly in the pilosebaceous follicle. P. acnes triggers a strong immune response involving keratinocytes, sebocytes and monocytes, the target cells during acne development. Lipoteicoic acid and peptidoglycan induce the inflammatory reaction, but no P. acnes surface protein interacting with Toll-like receptors has been identified. P. acnes surface proteins have been extracted by lithium stripping and shown to induce CXCL8 production by keratinocytes.
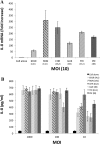
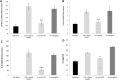
Methodology and principal findings: Far-western blotting identified two surface proteins, of 24.5- and 27.5-kDa in size, specifically recognized by TLR2. These proteins were characterized, by LC-MS/MS, as CAMP factor 1 devoid of its signal peptide sequence, as shown by N-terminal sequencing. Purified CAMP factor 1 induces CXCL8 production by activating the CXCL8 gene promoter, triggering the synthesis of CXCL8 mRNA. Antibodies against TLR2 significantly decreased the CXCL8 response. For the 27 P. acnes strains used in this study, CAMP1-TLR2 binding intensity was modulated and appeared to be strong in type IB and II strains, which produced large amounts of CXCL8, whereas most of the type IA1 and IA2 strains presented little or no CAMP1-TLR2 binding and low levels of CXCL8 production. The nucleotide sequence of CAMP factor displays a major polymorphism, defining two distinct genetic groups corresponding to CAMP factor 1 with 14 amino-acid changes from strains phylotyped II with moderate and high levels of CAMP1-TLR2 binding activity, and CAMP factor 1 containing 0, 1 or 2 amino-acid changes from strains phylotyped IA1, IA2, or IB presenting no, weak or moderate CAMP1-TLR2 binding.
Conclusions: Our findings indicate that CAMP factor 1 may contribute to P. acnes virulence, by amplifying the inflammation reaction through direct interaction with TLR2.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Activation of toll-like receptor 2 in acne triggers inflammatory cytokine responses.J Immunol. 2002 Aug 1;169(3):1535-41. doi: 10.4049/jimmunol.169.3.1535. J Immunol. 2002. PMID: 12133981 Free PMC article.
-
Distinct strains of Propionibacterium acnes induce selective human beta-defensin-2 and interleukin-8 expression in human keratinocytes through toll-like receptors.J Invest Dermatol. 2005 May;124(5):931-8. doi: 10.1111/j.0022-202X.2005.23705.x. J Invest Dermatol. 2005. PMID: 15854033
-
Review of the innate immune response in acne vulgaris: activation of Toll-like receptor 2 in acne triggers inflammatory cytokine responses.Dermatology. 2005;211(3):193-8. doi: 10.1159/000087011. Dermatology. 2005. PMID: 16205063 Review.
-
Characterization of a Cutibacterium acnes Camp Factor 1-Related Peptide as a New TLR-2 Modulator in In Vitro and Ex Vivo Models of Inflammation.Int J Mol Sci. 2022 May 3;23(9):5065. doi: 10.3390/ijms23095065. Int J Mol Sci. 2022. PMID: 35563458 Free PMC article.
-
Propionibacterium acnes: an update on its role in the pathogenesis of acne.J Eur Acad Dermatol Venereol. 2014 Mar;28(3):271-8. doi: 10.1111/jdv.12224. Epub 2013 Aug 1. J Eur Acad Dermatol Venereol. 2014. PMID: 23905540 Review.
Cited by
-
Antimicrobial production by perifollicular dermal preadipocytes is essential to the pathophysiology of acne.Sci Transl Med. 2022 Feb 16;14(632):eabh1478. doi: 10.1126/scitranslmed.abh1478. Epub 2022 Feb 16. Sci Transl Med. 2022. PMID: 35171653 Free PMC article.
-
In Vitro and Clinical Evaluation of Cannabigerol (CBG) Produced via Yeast Biosynthesis: A Cannabinoid with a Broad Range of Anti-Inflammatory and Skin Health-Boosting Properties.Molecules. 2022 Jan 13;27(2):491. doi: 10.3390/molecules27020491. Molecules. 2022. PMID: 35056807 Free PMC article.
-
A New Topical Candidate in Acne Treatment: Characterization of the Meclozine Hydrochloride as an Anti-Inflammatory Compound from In Vitro to a Preliminary Clinical Study.Biomedicines. 2022 Apr 19;10(5):931. doi: 10.3390/biomedicines10050931. Biomedicines. 2022. PMID: 35625668 Free PMC article.
-
Clinical and Biological Features of Cutibacterium (Formerly Propionibacterium) avidum, an Underrecognized Microorganism.Clin Microbiol Rev. 2018 May 30;31(3):e00064-17. doi: 10.1128/CMR.00064-17. Print 2018 Jul. Clin Microbiol Rev. 2018. PMID: 29848774 Free PMC article. Review.
-
Antibiotic Susceptibility of Cutibacterium acnes Strains Isolated from Israeli Acne Patients.Acta Derm Venereol. 2020 Oct 20;100(17):adv00295. doi: 10.2340/00015555-3654. Acta Derm Venereol. 2020. PMID: 33021324 Free PMC article.
References
-
- Aubin GG, Portillo ME, Trampuz A, Corvec S. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Medecine Mal Infect. 2014; 44: 241–250. - PubMed
-
- Lalani T, Person AK, Hedayati SS, Moore L, Murdoch DR, Hoen B, et al. (2007). Propionibacterium endocarditis: a case series from the International Collaboration on Endocarditis Merged Database and Prospective Cohort Study. Scand J Infect Dis. 2007; 39: 840–848. 10.1080/00365540701367793 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

