Determinants of tRNA Recognition by the Radical SAM Enzyme RlmN
- PMID: 27902775
- PMCID: PMC5130265
- DOI: 10.1371/journal.pone.0167298
Determinants of tRNA Recognition by the Radical SAM Enzyme RlmN
Abstract
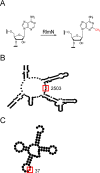
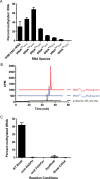
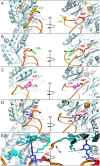
RlmN, a bacterial radical SAM methylating enzyme, has the unusual ability to modify two distinct types of RNA: 23S rRNA and tRNA. In rRNA, RlmN installs a methyl group at the C2 position of A2503 of 23S rRNA, while in tRNA the modification occurs at nucleotide A37, immediately adjacent to the anticodon triplet. Intriguingly, only a subset of tRNAs that contain an adenosine at position 37 are substrates for RlmN, suggesting that the enzyme carefully probes the highly conserved tRNA fold and sequence features to identify its targets. Over the past several years, multiple studies have addressed rRNA modification by RlmN, while relatively few investigations have focused on the ability of this enzyme to modify tRNAs. In this study, we utilized in vitro transcribed tRNAs as model substrates to interrogate RNA recognition by RlmN. Using chimeras and point mutations, we probed how the structure and sequence of RNA influences methylation, identifying position 38 of tRNAs as a critical determinant of substrate recognition. We further demonstrate that, analogous to previous mechanistic studies with fragments of 23S rRNA, tRNA methylation requirements are consistent with radical SAM reactivity. Together, our findings provide detailed insight into tRNA recognition by a radical SAM methylating enzyme.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy.RNA. 2012 Oct;18(10):1783-95. doi: 10.1261/rna.033266.112. Epub 2012 Aug 13. RNA. 2012. PMID: 22891362 Free PMC article.
-
A QM/MM study of the catalytic mechanism of SAM methyltransferase RlmN from Escherichia coli.Proteins. 2017 Nov;85(11):1967-1974. doi: 10.1002/prot.25337. Epub 2017 Aug 29. Proteins. 2017. PMID: 28643349
-
Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating enzyme.Nucleic Acids Res. 2016 Oct 14;44(18):8897-8907. doi: 10.1093/nar/gkw699. Epub 2016 Aug 5. Nucleic Acids Res. 2016. PMID: 27496281 Free PMC article.
-
Radical-mediated enzymatic methylation: a tale of two SAMS.Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review.
-
Radical SAM-mediated methylation reactions.Curr Opin Chem Biol. 2013 Aug;17(4):597-604. doi: 10.1016/j.cbpa.2013.05.032. Epub 2013 Jul 5. Curr Opin Chem Biol. 2013. PMID: 23835516 Free PMC article. Review.
Cited by
-
The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function.Nucleic Acids Res. 2019 Mar 18;47(5):2446-2454. doi: 10.1093/nar/gkz030. Nucleic Acids Res. 2019. PMID: 30698741 Free PMC article.
-
New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1.RNA Biol. 2021 Dec;18(12):2531-2545. doi: 10.1080/15476286.2021.1930756. Epub 2021 Jun 10. RNA Biol. 2021. PMID: 34110975 Free PMC article.
-
miCLIP-MaPseq, a Substrate Identification Approach for Radical SAM RNA Methylating Enzymes.J Am Chem Soc. 2018 Jun 13;140(23):7135-7143. doi: 10.1021/jacs.8b02618. Epub 2018 Jun 5. J Am Chem Soc. 2018. PMID: 29782154 Free PMC article.
-
miCLIP-MaPseq Identifies Substrates of Radical SAM RNA-Methylating Enzyme Using Mechanistic Cross-Linking and Mismatch Profiling.Methods Mol Biol. 2021;2298:105-122. doi: 10.1007/978-1-0716-1374-0_7. Methods Mol Biol. 2021. PMID: 34085241 Free PMC article.
-
The modification landscape of Pseudomonas aeruginosa tRNAs.RNA. 2024 Jul 16;30(8):1025-1040. doi: 10.1261/rna.080004.124. RNA. 2024. PMID: 38684317 Free PMC article.
References
-
- Golovina AY, Dzama MM, Osterman IA, Sergiev PV, Serebryakova MV, Bogdanov AA, et al. The last rRNA methyltransferase of E. coli revealed: The yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA. 2012; 18: 1725–1734. 10.1261/rna.034207.112 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

