RAC1 GTP-ase signals Wnt-beta-catenin pathway mediated integrin-directed metastasis-associated tumor cell phenotypes in triple negative breast cancers
- PMID: 27902969
- PMCID: PMC5356866
- DOI: 10.18632/oncotarget.13618
RAC1 GTP-ase signals Wnt-beta-catenin pathway mediated integrin-directed metastasis-associated tumor cell phenotypes in triple negative breast cancers
Abstract
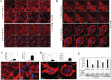
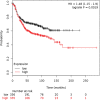
The acquisition of integrin-directed metastasis-associated (ID-MA) phenotypes by Triple-Negative Breast Cancer (TNBC) cells is caused by an upregulation of the Wnt-beta-catenin pathway (WP). We reported that WP is one of the salient genetic features of TNBC. RAC-GTPases, small G-proteins which transduce signals from cell surface proteins including integrins, have been implicated in tumorigenesis and metastasis by their role in essential cellular functions like motility. The collective percentage of alteration(s) in RAC1 in ER+ve BC was lower as compared to ER-ve BC (35% vs 57%) (brca/tcga/pub2015). High expression of RAC1 was associated with poor outcome for RFS with HR=1.48 [CI: 1.15-1.9] p=0.0019 in the Hungarian ER-veBC cohort. Here we examined how WP signals are transduced via RAC1 in the context of ID-MA phenotypes in TNBC. Using pharmacological agents (sulindac sulfide), genetic tools (beta-catenin siRNA), WP modulators (Wnt-C59, XAV939), RAC1 inhibitors (NSC23766, W56) and WP stimulations (LWnt3ACM, Wnt3A recombinant) in a panel of 6-7 TNBC cell lines, we studied fibronectin-directed (1) migration, (2) matrigel invasion, (3) RAC1 and Cdc42 activation, (4) actin dynamics (confocal microscopy) and (5) podia-parameters. An attenuation of WP, which (a) decreased cellular levels of beta-catenin, as well as its nuclear active-form, (b) decreased fibronectin-induced migration, (c) decreased invasion, (d) altered actin dynamics and (e) decreased podia-parameters was successful in blocking fibronectin-mediated RAC1/Cdc42 activity. Both Wnt-antagonists and RAC1 inhibitors blocked fibronectin-induced RAC1 activation and inhibited the fibronectin-induced ID-MA phenotypes following specific WP stimulation by LWnt3ACM as well as Wnt3A recombinant protein. To test a direct involvement of RAC1-activation in WP-mediated ID-MA phenotypes, we stimulated brain-metastasis specific MDA-MB231BR cells with LWnt3ACM. LWnt3ACM-stimulated fibronectin-directed migration was blocked by RAC1 inhibition in MDA-MB231BR cells. In the light of our previous report that WP upregulation causes ID-MA phenotypes in TNBC tumor cells, here we provide the first mechanism based evidence to demonstrate that WP upregulation signals ID-MA tumor cell phenotypes in a RAC1-GTPase dependent manner involving exchange-factors like TIAM1 and VAV2. Our study demonstrates for the first time that beta-catenin-RAC1 cascade signals integrin-directed metastasis-associated tumor cell phenotypes in TNBC.
Keywords: RAC1 GTP-ase; Wnt pathway; actin cytoskeleton; brain-metastasis specific cells; integrin-directed migration.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
-
- Entschladen F, TLt Drell, Lang K, Joseph J, Zaenker KS. Tumour-cell migration, invasion, and metastasis: navigation by neurotransmitters. The Lancet Oncology. 2004;5:254–258. - PubMed
-
- Lorusso G, Ruegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochemistry and cell biology. 2008;130:1091–1103. - PubMed
-
- Cavallaro U, Christofori G. Cell adhesion in tumor invasion and metastasis: loss of the glue is not enough. Biochimica et biophysica acta. 2001;1552:39–45. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

