Synthetic biology routes to bio-artificial intelligence
- PMID: 27903825
- PMCID: PMC5264507
- DOI: 10.1042/EBC20160014
Synthetic biology routes to bio-artificial intelligence
Abstract
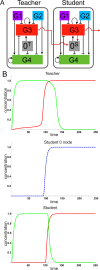
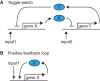
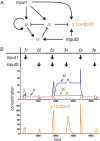
The design of synthetic gene networks (SGNs) has advanced to the extent that novel genetic circuits are now being tested for their ability to recapitulate archetypal learning behaviours first defined in the fields of machine and animal learning. Here, we discuss the biological implementation of a perceptron algorithm for linear classification of input data. An expansion of this biological design that encompasses cellular 'teachers' and 'students' is also examined. We also discuss implementation of Pavlovian associative learning using SGNs and present an example of such a scheme and in silico simulation of its performance. In addition to designed SGNs, we also consider the option to establish conditions in which a population of SGNs can evolve diversity in order to better contend with complex input data. Finally, we compare recent ethical concerns in the field of artificial intelligence (AI) and the future challenges raised by bio-artificial intelligence (BI).
Keywords: artificial intelligence; gene networks; synthetic biological circuits; synthetic biology.
© 2016 The Author(s). This is an open access article published by Portland Press Limited on behalf of the Biochemical Society and distributed under the Creative Commons Attribution License 4.0 (CC BY).
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

