Elasticity of the transition state for oligonucleotide hybridization
- PMID: 27903889
- PMCID: PMC5314771
- DOI: 10.1093/nar/gkw1173
Elasticity of the transition state for oligonucleotide hybridization
Abstract
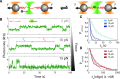
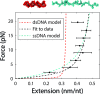
Despite its fundamental importance in cellular processes and abundant use in biotechnology, we lack a detailed understanding of the kinetics of nucleic acid hybridization. In particular, the identity of the transition state, which determines the kinetics of the two-state reaction, remains poorly characterized. Here, we used optical tweezers with single-molecule fluorescence to observe directly the binding and unbinding of short oligonucleotides (7-12 nt) to a complementary strand held under constant force. Binding and unbinding rate constants measured across a wide range of forces (1.5-20 pN) deviate from the exponential force dependence expected from Bell's equation. Using a generalized force dependence model, we determined the elastic behavior of the transition state, which we find to be similar to that of the pure single-stranded state. Our results indicate that the transition state for hybridization is visited before the strands form any significant amount of native base pairs. Such a transition state supports a model in which the rate-limiting step of the hybridization reaction is the alignment of the two strands prior to base pairing.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Erlich H.A., Gelfand D., Sninsky J.J. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. - PubMed
-
- Dorsett Y., Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. - PubMed
-
- Yurke B., Turber A.J., Mills A.P., Jr, Simmel F.C., Neumann J.L. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. - PubMed
-
- Sherman W.B., Seeman N.C. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203–1207.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

