Autoregulation of MBNL1 function by exon 1 exclusion from MBNL1 transcript
- PMID: 27903900
- PMCID: PMC5389549
- DOI: 10.1093/nar/gkw1158
Autoregulation of MBNL1 function by exon 1 exclusion from MBNL1 transcript
Abstract
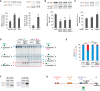
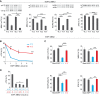
Muscleblind-like proteins (MBNLs) are regulators of RNA metabolism. During tissue differentiation the level of MBNLs increases, while their functional insufficiency plays a crucial role in myotonic dystrophy (DM). Deep sequencing of RNA molecules cross-linked to immunoprecipitated protein particles (CLIP-seq) revealed that MBNL1 binds to MBNL1 exon 1 (e1) encoding both the major part of 5΄UTR and an amino-terminal region of MBNL1 protein. We tested several hypotheses regarding the possible autoregulatory function of MBNL1 binding to its own transcript. Our data indicate that MBNLs induce skipping of e1 from precursor MBNL1 mRNA and that e1 exclusion may impact transcript association with polysomes and translation. Furthermore, e1-deficient protein isoform lacking the first two zinc fingers is highly unstable and its EGFP fusion protein has severely compromised splicing activity. We also show that MBNL1 can be transcribed from three different promoters and that the transcription initiation site determines the mode of e1 regulation. Taken together, we demonstrate that MBNL proteins control steady-state levels of MBNL1 through an interaction with e1 in its precursor mRNA. Insights from our study open a new avenue in therapies against DM based on manipulation of the transcription initiation site and e1 splicing of MBNL1 mRNA.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
MBNL expression in autoregulatory feedback loops.RNA Biol. 2018 Jan 2;15(1):1-8. doi: 10.1080/15476286.2017.1384119. Epub 2017 Nov 13. RNA Biol. 2018. PMID: 28949831 Free PMC article.
-
Conserved developmental alternative splicing of muscleblind-like (MBNL) transcripts regulates MBNL localization and activity.RNA Biol. 2010 Jan-Feb;7(1):43-55. doi: 10.4161/rna.7.1.10401. Epub 2010 Jan 21. RNA Biol. 2010. PMID: 20009516
-
Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs.Nucleic Acids Res. 2007;35(16):5474-86. doi: 10.1093/nar/gkm601. Epub 2007 Aug 15. Nucleic Acids Res. 2007. PMID: 17702765 Free PMC article.
-
The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing.Differentiation. 2006 Mar;74(2-3):65-80. doi: 10.1111/j.1432-0436.2006.00060.x. Differentiation. 2006. PMID: 16533306 Review.
-
MBNL proteins and their target RNAs, interaction and splicing regulation.Nucleic Acids Res. 2014;42(17):10873-87. doi: 10.1093/nar/gku767. Epub 2014 Sep 2. Nucleic Acids Res. 2014. PMID: 25183524 Free PMC article. Review.
Cited by
-
Transcriptome regulation by PARP13 in basal and antiviral states in human cells.iScience. 2024 Feb 16;27(4):109251. doi: 10.1016/j.isci.2024.109251. eCollection 2024 Apr 19. iScience. 2024. PMID: 38495826 Free PMC article.
-
Alternative splicing and related RNA binding proteins in human health and disease.Signal Transduct Target Ther. 2024 Feb 2;9(1):26. doi: 10.1038/s41392-024-01734-2. Signal Transduct Target Ther. 2024. PMID: 38302461 Free PMC article. Review.
-
The X-linked splicing regulator MBNL3 has been co-opted to restrict placental growth in eutherians.PLoS Biol. 2022 Apr 27;20(4):e3001615. doi: 10.1371/journal.pbio.3001615. eCollection 2022 Apr. PLoS Biol. 2022. PMID: 35476669 Free PMC article.
-
(CTG)n repeat-mediated dysregulation of MBNL1 and MBNL2 expression during myogenesis in DM1 occurs already at the myoblast stage.PLoS One. 2019 May 22;14(5):e0217317. doi: 10.1371/journal.pone.0217317. eCollection 2019. PLoS One. 2019. PMID: 31116797 Free PMC article.
-
Myotonic dystrophy RNA toxicity alters morphology, adhesion and migration of mouse and human astrocytes.Nat Commun. 2022 Jul 4;13(1):3841. doi: 10.1038/s41467-022-31594-9. Nat Commun. 2022. PMID: 35789154 Free PMC article.
References
-
- Rau F., Freyermuth F., Fugier C., Villemin J.-P., Fischer M.-C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D. et al. . Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011; 18:840–845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

