Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation
- PMID: 27903902
- PMCID: PMC5389550
- DOI: 10.1093/nar/gkw1163
Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation
Abstract
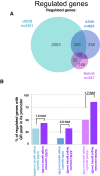
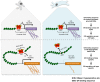
The genomic loci bound by the glucocorticoid receptor (GR), a hormone-activated transcription factor, show little overlap between cell types. To study the role of chromatin and sequence in specifying where GR binds, we used Bayesian modeling within the universe of accessible chromatin. Taken together, our results uncovered that although GR preferentially binds accessible chromatin, its binding is biased against accessible chromatin located at promoter regions. This bias can only be explained partially by the presence of fewer GR recognition sequences, arguing for the existence of additional mechanisms that interfere with GR binding at promoters. Therefore, we tested the role of H3K9ac, the chromatin feature with the strongest negative association with GR binding, but found that this correlation does not reflect a causative link. Finally, we find a higher percentage of promoter-proximal GR binding for genes regulated by GR across cell types than for cell type-specific target genes. Given that GR almost exclusively binds accessible chromatin, we propose that cell type-specific regulation by GR preferentially occurs via distal enhancers, whose chromatin accessibility is typically cell type-specific, whereas ubiquitous target gene regulation is more likely to result from binding to promoter regions, which are often accessible regardless of cell type examined.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Siersbaek R., Rabiee A., Nielsen R., Sidoli S., Traynor S., Loft A., La Cour Poulsen L., Rogowska-Wrzesinska A., Jensen O.N., Mandrup S.. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014; 7:1443–1455. - PubMed
-
- Jolma A., Yin Y., Nitta K.R., Dave K., Popov A., Taipale M., Enge M., Kivioja T., Morgunova E., Taipale J.. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015; 527:384–388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

