Structural insights into dynamics of RecU-HJ complex formation elucidates key role of NTR and stalk region toward formation of reactive state
- PMID: 27903910
- PMCID: PMC5314769
- DOI: 10.1093/nar/gkw1165
Structural insights into dynamics of RecU-HJ complex formation elucidates key role of NTR and stalk region toward formation of reactive state
Abstract
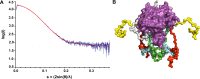
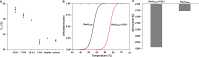
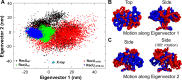
Holliday junction (HJ) resolving enzyme RecU is involved in DNA repair and recombination. We have determined the crystal structure of inactive mutant (D88N) of RecU from Bacillus subtilis in complex with a 12 base palindromic DNA fragment at a resolution of 3.2 Å. This structure shows the stalk region and the essential N-terminal region (NTR) previously unseen in our DNA unbound structure. The flexible nature of the NTR in solution was confirmed using SAXS. Thermofluor studies performed to assess the stability of RecU in complex with the arms of an HJ indicate that it confers stability. Further, we performed molecular dynamics (MD) simulations of wild type and an NTR deletion variant of RecU, with and without HJ. The NTR is observed to be highly flexible in simulations of the unbound RecU, in agreement with SAXS observations. These simulations revealed domain dynamics of RecU and their role in the formation of complex with HJ. The MD simulations also elucidate key roles of the NTR, stalk region, and breathing motion of RecU in the formation of the reactive state.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Direct analysis of Holliday junction resolving enzyme in a DNA origami nanostructure.Nucleic Acids Res. 2014 Jun;42(11):7421-8. doi: 10.1093/nar/gku320. Epub 2014 May 3. Nucleic Acids Res. 2014. PMID: 24792171 Free PMC article.
-
Identification of amino acid residues critical for catalysis of Holliday junction resolution by Mycoplasma genitalium RecU.J Bacteriol. 2011 Aug;193(15):3941-8. doi: 10.1128/JB.00247-11. Epub 2011 Jun 3. J Bacteriol. 2011. PMID: 21642467 Free PMC article.
-
The stalk region of the RecU resolvase is essential for Holliday junction recognition and distortion.J Mol Biol. 2011 Jul 1;410(1):39-49. doi: 10.1016/j.jmb.2011.05.008. Epub 2011 May 12. J Mol Biol. 2011. PMID: 21600217
-
Holliday junction resolvases.Cold Spring Harb Perspect Biol. 2014 Sep 2;6(9):a023192. doi: 10.1101/cshperspect.a023192. Cold Spring Harb Perspect Biol. 2014. PMID: 25183833 Free PMC article. Review.
-
New insight into the recognition of branched DNA structure by junction-resolving enzymes.Curr Opin Struct Biol. 2008 Feb;18(1):86-95. doi: 10.1016/j.sbi.2007.11.001. Epub 2007 Dec 21. Curr Opin Struct Biol. 2008. PMID: 18160275 Review.
Cited by
-
RecA Regulation by RecU and DprA During Bacillus subtilis Natural Plasmid Transformation.Front Microbiol. 2018 Jul 11;9:1514. doi: 10.3389/fmicb.2018.01514. eCollection 2018. Front Microbiol. 2018. PMID: 30050509 Free PMC article.
-
DisA Restrains the Processing and Cleavage of Reversed Replication Forks by the RuvAB-RecU Resolvasome.Int J Mol Sci. 2021 Oct 20;22(21):11323. doi: 10.3390/ijms222111323. Int J Mol Sci. 2021. PMID: 34768753 Free PMC article.
-
Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.Sci Rep. 2020 Jan 15;10(1):393. doi: 10.1038/s41598-019-56825-w. Sci Rep. 2020. PMID: 31941902 Free PMC article.
-
Wordom update 2: A user-friendly program for the analysis of molecular structures and conformational ensembles.Comput Struct Biotechnol J. 2023 Jan 27;21:1390-1402. doi: 10.1016/j.csbj.2023.01.026. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36817953 Free PMC article.
-
The phage protein paratox is a multifunctional metabolic regulator of Streptococcus.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1200. doi: 10.1093/nar/gkae1200. Nucleic Acids Res. 2025. PMID: 39673798 Free PMC article.
References
-
- West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. - PubMed
-
- Lloyd K.B., Low R.G. Homologous recombination. In: Neidhardt FC, Curtiss R III, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, D.C.: ASM Press; 1996. pp. 2236–2255.
-
- Cañas C., Carrasco B., García-Tirado E., Rafferty J.B., Alonso J.C., Ayora S. The stalk region of the RecU resolvase is essential for Holliday junction recognition and distortion. J. Mol. Biol. 2011;410:39–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

