miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA-target interaction data
- PMID: 27903911
- PMCID: PMC5389546
- DOI: 10.1093/nar/gkw1185
miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA-target interaction data
Abstract
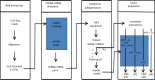
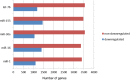
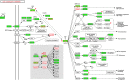
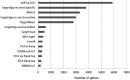
MicroRNAs (miRNAs) are ∼19-22 nucleotides (nt) long regulatory RNAs that regulate gene expression by recognizing and binding to complementary sequences on mRNAs. The key step in revealing the function of a miRNA, is the identification of miRNA target genes. Recent biochemical advances including PAR-CLIP and HITS-CLIP allow for improved miRNA target predictions and are widely used to validate miRNA targets. Here, we present miRTar2GO, which is a model, trained on the common rules of miRNA-target interactions, Argonaute (Ago) CLIP-Seq data and experimentally validated miRNA target interactions. miRTar2GO is designed to predict miRNA target sites using more relaxed miRNA-target binding characteristics. More importantly, miRTar2GO allows for the prediction of cell-type specific miRNA targets. We have evaluated miRTar2GO against other widely used miRNA target prediction algorithms and demonstrated that miRTar2GO produced significantly higher F1 and G scores. Target predictions, binding specifications, results of the pathway analysis and gene ontology enrichment of miRNA targets are freely available at http://www.mirtar2go.org.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Tran N., Hutvagner G.. Biogenesis and the regulation of the maturation of miRNAs. Essays Biochem. 2013; 54:17–28. - PubMed
-
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. - PubMed
-
- Rivas F. V, Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L.. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005; 12:340–349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

