Microbial Community Patterns Associated with Automated Teller Machine Keypads in New York City
- PMID: 27904880
- PMCID: PMC5112336
- DOI: 10.1128/mSphere.00226-16
Microbial Community Patterns Associated with Automated Teller Machine Keypads in New York City
Abstract
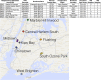
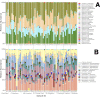
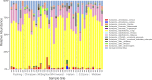
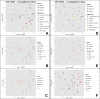
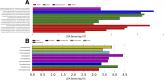
In densely populated urban environments, the distribution of microbes and the drivers of microbial community assemblages are not well understood. In sprawling metropolitan habitats, the "urban microbiome" may represent a mix of human-associated and environmental taxa. Here we carried out a baseline study of automated teller machine (ATM) keypads in New York City (NYC). Our goal was to describe the biodiversity and biogeography of both prokaryotic and eukaryotic microbes in an urban setting while assessing the potential source of microbial assemblages on ATM keypads. Microbial swab samples were collected from three boroughs (Manhattan, Queens, and Brooklyn) during June and July 2014, followed by generation of Illumina MiSeq datasets for bacterial (16S rRNA) and eukaryotic (18S rRNA) marker genes. Downstream analysis was carried out in the QIIME pipeline, in conjunction with neighborhood metadata (ethnicity, population, age groups) from the NYC Open Data portal. Neither the 16S nor 18S rRNA datasets showed any clustering patterns related to geography or neighborhood demographics. Bacterial assemblages on ATM keypads were dominated by taxonomic groups known to be associated with human skin communities (Actinobacteria, Bacteroides, Firmicutes, and Proteobacteria), although SourceTracker analysis was unable to identify the source habitat for the majority of taxa. Eukaryotic assemblages were dominated by fungal taxa as well as by a low-diversity protist community containing both free-living and potentially pathogenic taxa (Toxoplasma, Trichomonas). Our results suggest that ATM keypads amalgamate microbial assemblages from different sources, including the human microbiome, eukaryotic food species, and potentially novel extremophilic taxa adapted to air or surfaces in the built environment. DNA obtained from ATM keypads may thus provide a record of both human behavior and environmental sources of microbes. IMPORTANCE Automated teller machine (ATM) keypads represent a specific and unexplored microhabitat for microbial communities. Although the number of built environment and urban microbial ecology studies has expanded greatly in recent years, the majority of research to date has focused on mass transit systems, city soils, and plumbing and ventilation systems in buildings. ATM surfaces, potentially retaining microbial signatures of human inhabitants, including both commensal taxa and pathogens, are interesting from both a biodiversity perspective and a public health perspective. By focusing on ATM keypads in different geographic areas of New York City with distinct population demographics, we aimed to characterize the diversity and distribution of both prokaryotic and eukaryotic microbes, thus making a unique contribution to the growing body of work focused on the "urban microbiome." In New York City, the surface area of urban surfaces in Manhattan far exceeds the geographic area of the island itself. We have only just begun to describe the vast array of microbial taxa that are likely to be present across diverse types of urban habitats.
Keywords: 16S rRNA; 18S rRNA; New York City; automated teller machine; environmental sequencing; urban microbiome.
Figures

References
-
- Creer S, Deiner K, Frey S, Porazinska D, Taberlet P, Thomas WK, Potter C, Bik HM. 2016. The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol Evol 7:1008–1018. doi: 10.1111/2041-210X.12574. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

