The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity
- PMID: 27905489
- PMCID: PMC5131312
- DOI: 10.1038/srep38063
The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity
Abstract
Mechanical integration of the nucleus with the extracellular matrix (ECM) is established by linkage between the cytoskeleton and the nucleus. This integration is hypothesized to mediate sensing of ECM rigidity, but parsing the function of nucleus-cytoskeleton linkage from other mechanisms has remained a central challenge. Here we took advantage of the fact that the LINC (linker of nucleoskeleton and cytoskeleton) complex is a known molecular linker of the nucleus to the cytoskeleton, and asked how it regulates the sensitivity of genome-wide transcription to substratum rigidity. We show that gene mechanosensitivity is preserved after LINC disruption, but reversed in direction. Combined with myosin inhibition studies, we identify genes that depend on nuclear tension for their regulation. We also show that LINC disruption does not attenuate nuclear shape sensitivity to substrate rigidity. Our results show for the first time that the LINC complex facilitates mechano-regulation of expression across the genome.
Figures

 , blue solid circles), other genes lost mechanosensitivity (
, blue solid circles), other genes lost mechanosensitivity ( , yellow solid circles) and the majority of genes reversed the direction of mechanosensitivity (
, yellow solid circles) and the majority of genes reversed the direction of mechanosensitivity ( , red solid circles). Mechanosensitivity is calculated as the ratio of the expression levels (EL) between the two substrate rigidities as shown.
, red solid circles). Mechanosensitivity is calculated as the ratio of the expression levels (EL) between the two substrate rigidities as shown.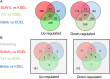

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

