Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida
- PMID: 27905529
- PMCID: PMC5131344
- DOI: 10.1038/srep38169
Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida
Abstract
CRISPR/Cas9 systems are nowadays applied extensively to effect genome editing in various organisms including plants. CRISPR from Prevotella and Francisella 1 (Cpf1) is a newly characterized RNA-guided endonuclease that has two distinct features as compared to Cas9. First, Cpf1 utilizes a thymidine-rich protospacer adjacent motif (PAM) while Cas9 prefers a guanidine-rich PAM. Cpf1 could be used as a sequence-specific nuclease to target AT-rich regions of a genome that Cas9 had difficulty accessing. Second, Cpf1 generates DNA ends with a 5' overhang, whereas Cas9 creates blunt DNA ends after cleavage. "Sticky" DNA ends should increase the efficiency of insertion of a desired DNA fragment into the Cpf1-cleaved site using complementary DNA ends. Therefore, Cpf1 could be a potent tool for precise genome engineering. To evaluate whether Cpf1 can be applied to plant genome editing, we selected Cpf1 from Francisella novicida (FnCpf1), which recognizes a shorter PAM (TTN) within known Cpf1 proteins, and applied it to targeted mutagenesis in tobacco and rice. Our results show that targeted mutagenesis had occurred in transgenic plants expressing FnCpf1 with crRNA. Deletions of the targeted region were the most frequently observed mutations. Our results demonstrate that FnCpf1 can be applied successfully to genome engineering in plants.
Figures
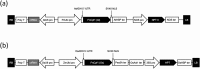

References
-
- Osakabe Y. & Osakabe K. Genome editing with engineered nucleases in plants. Plant Cell Physiol. 56, 389–400 (2015). - PubMed
-
- Voytas D. F. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350 (2013). - PubMed
-
- Kumar V. & Jain M. The CRISPR-Cas system for plant genome editing: advances and opportunities. J. Exp. Bot. 66, 47–57 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

