Deep, Staged Transcriptomic Resources for the Novel Coleopteran Models Atrachya menetriesi and Callosobruchus maculatus
- PMID: 27907180
- PMCID: PMC5132259
- DOI: 10.1371/journal.pone.0167431
Deep, Staged Transcriptomic Resources for the Novel Coleopteran Models Atrachya menetriesi and Callosobruchus maculatus
Abstract
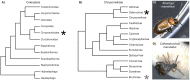
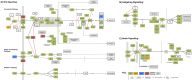
Despite recent efforts to sample broadly across metazoan and insect diversity, current sequence resources in the Coleoptera do not adequately describe the diversity of the clade. Here we present deep, staged transcriptomic data for two coleopteran species, Atrachya menetriesi (Faldermann 1835) and Callosobruchus maculatus (Fabricius 1775). Our sampling covered key stages in ovary and early embryonic development in each species. We utilized this data to build combined assemblies for each species which were then analysed in detail. The combined A. menetriesi assembly consists of 228,096 contigs with an N50 of 1,598 bp, while the combined C. maculatus assembly consists of 128,837 contigs with an N50 of 2,263 bp. For these assemblies, 34.6% and 32.4% of contigs were identified using Blast2GO, and 97% and 98.3% of the BUSCO set of metazoan orthologs were present, respectively. We also carried out manual annotation of developmental signalling pathways and found that nearly all expected genes were present in each transcriptome. Our analyses show that both transcriptomes are of high quality. Lastly, we performed read mapping utilising our timed, stage specific RNA samples to identify differentially expressed contigs. The resources presented here will provide a firm basis for a variety of experimentation, both in developmental biology and in comparative genomic studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Stork NE. Insect diversity—facts, fiction and speculation. Biol J Linn Soc. 24–28 OVAL RD, LONDON, ENGLAND NW1 7DX: ACADEMIC PRESS LTD; 1988;35: 321–337.
-
- Grimaldi D, Engel M. Evolution of the Insects. Cambridge University Press; 2005.
-
- 1KITE: 1000 Insect Transcriptome Evolution [Internet]. 2015. http://www.1kite.org/
-
- Keeling CI, Henderson H, Li M, Yuen M, Clark EL, Fraser JD, et al. Transcriptome and full-length cDNA resources for the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major insect pest of pine forests. Insect Biochem Mol Biol. England; 2012;42: 525–536. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

