Genome-Editing Technologies: Principles and Applications
- PMID: 27908936
- PMCID: PMC5131771
- DOI: 10.1101/cshperspect.a023754
Genome-Editing Technologies: Principles and Applications
Abstract
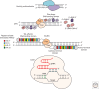
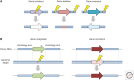
Targeted nucleases have provided researchers with the ability to manipulate virtually any genomic sequence, enabling the facile creation of isogenic cell lines and animal models for the study of human disease, and promoting exciting new possibilities for human gene therapy. Here we review three foundational technologies-clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9), transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases (ZFNs). We discuss the engineering advances that facilitated their development and highlight several achievements in genome engineering that were made possible by these tools. We also consider artificial transcription factors, illustrating how this technology can complement targeted nucleases for synthetic biology and gene therapy.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
References
-
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. 2003. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol 21: 275–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
