The small FOXP1 isoform predominantly expressed in activated B cell-like diffuse large B-cell lymphoma and full-length FOXP1 exert similar oncogenic and transcriptional activity in human B cells
- PMID: 27909217
- PMCID: PMC5394978
- DOI: 10.3324/haematol.2016.156455
The small FOXP1 isoform predominantly expressed in activated B cell-like diffuse large B-cell lymphoma and full-length FOXP1 exert similar oncogenic and transcriptional activity in human B cells
Abstract
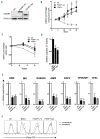
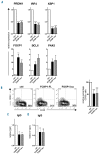
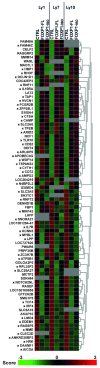
The forkhead transcription factor FOXP1 is generally regarded as an oncogene in activated B cell-like diffuse large B-cell lymphoma. Previous studies have suggested that a small isoform of FOXP1 rather than full-length FOXP1, may possess this oncogenic activity. Corroborating those studies, we herein show that activated B cell-like diffuse large B-cell lymphoma cell lines and primary activated B cell-like diffuse large B-cell lymphoma cells predominantly express a small FOXP1 isoform, and that the 5'-end of the Foxp1 gene is a common insertion site in murine lymphomas in leukemia virus- and transposon-mediated insertional mutagenesis screens. By combined mass spectrometry, (quantative) reverse transcription polymerase chain reaction/sequencing, and small interfering ribonucleic acid-mediated gene silencing, we determined that the small FOXP1 isoform predominantly expressed in activated B cell-like diffuse large B-cell lymphoma lacks the N-terminal 100 amino acids of full-length FOXP1. Aberrant overexpression of this FOXP1 isoform (ΔN100) in primary human B cells revealed its oncogenic capacity; it repressed apoptosis and plasma cell differentiation. However, no difference in potency was found between this small FOXP1 isoform and full-length FOXP1. Furthermore, overexpression of full-length FOXP1 or this small FOXP1 isoform in primary B cells and diffuse large B-cell lymphoma cell lines resulted in similar gene regulation. Taken together, our data indicate that this small FOXP1 isoform and full-length FOXP1 have comparable oncogenic and transcriptional activity in human B cells, suggesting that aberrant expression or overexpression of FOXP1, irrespective of the specific isoform, contributes to lymphomagenesis. These novel insights further enhance the value of FOXP1 for the diagnostics, prognostics, and treatment of diffuse large B-cell lymphoma patients.
Copyright© Ferrata Storti Foundation.
Figures


References
-
- Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328(2):198–206. - PubMed
-
- Zhang Y, Zhang S, Wang X, et al. Prognostic significance of FOXP1 as an oncogene in hepatocellular carcinoma. J Clin Pathol. 2012;65(6):528–533. - PubMed
-
- Rad R, Rad L, Wang W, et al. A conditional piggyBac transposition system for genetic screening in mice identifies oncogenic networks in pancreatic cancer. Nat Genet. 2015;47:47–56. - PubMed
-
- Banham AH, Connors JM, Brown PJ, et al. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2005;11(3): 1065–1072. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

