Genome-wide characterisation and expression profile of the grapevine ATL ubiquitin ligase family reveal biotic and abiotic stress-responsive and development-related members
- PMID: 27910910
- PMCID: PMC5133618
- DOI: 10.1038/srep38260
Genome-wide characterisation and expression profile of the grapevine ATL ubiquitin ligase family reveal biotic and abiotic stress-responsive and development-related members
Abstract
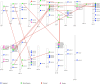
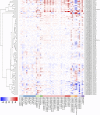
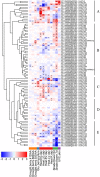
The Arabidopsis Tóxicos en Levadura (ATL) protein family is a class of E3 ubiquitin ligases with a characteristic RING-H2 Zn-finger structure that mediates diverse physiological processes and stress responses in plants. We carried out a genome-wide survey of grapevine (Vitis vinifera L.) ATL genes and retrieved 96 sequences containing the canonical ATL RING-H2 domain. We analysed their genomic organisation, gene structure and evolution, protein domains and phylogenetic relationships. Clustering revealed several clades, as already reported in Arabidopsis thaliana and rice (Oryza sativa), with an expanded subgroup of grapevine-specific genes. Most of the grapevine ATL genes lacked introns and were scattered among the 19 chromosomes, with a high level of duplication retention. Expression profiling revealed that some ATL genes are expressed specifically during early or late development and may participate in the juvenile to mature plant transition, whereas others may play a role in pathogen and/or abiotic stress responses, making them key candidates for further functional analysis. Our data offer the first genome-wide overview and annotation of the grapevine ATL family, and provide a basis for investigating the roles of specific family members in grapevine physiology and stress responses, as well as potential biotechnological applications.
Figures
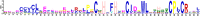



References
-
- Vierstra R. D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10, 385–397 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

