Transcription by RNA polymerase III: insights into mechanism and regulation
- PMID: 27911719
- PMCID: PMC5095917
- DOI: 10.1042/BST20160062
Transcription by RNA polymerase III: insights into mechanism and regulation
Abstract
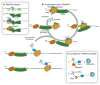
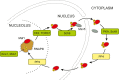
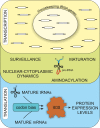
The highly abundant, small stable RNAs that are synthesized by RNA polymerase III (RNAPIII) have key functional roles, particularly in the protein synthesis apparatus. Their expression is metabolically demanding, and is therefore coupled to changing demands for protein synthesis during cell growth and division. Here, we review the regulatory mechanisms that control the levels of RNAPIII transcripts and discuss their potential physiological relevance. Recent analyses have revealed differential regulation of tRNA expression at all steps on its biogenesis, with significant deregulation of mature tRNAs in cancer cells.
Keywords: RNA polymerase III; tRNA; transcription elongation; transcription factors.
© 2016 The Author(s).
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

