Permissive roles of cytokines interleukin-7 and Flt3 ligand in mouse B-cell lineage commitment
- PMID: 27911806
- PMCID: PMC5167207
- DOI: 10.1073/pnas.1613316113
Permissive roles of cytokines interleukin-7 and Flt3 ligand in mouse B-cell lineage commitment
Abstract
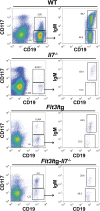
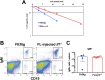
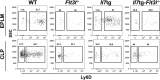
Hematopoietic cells are continuously generated throughout life from hematopoietic stem cells, thus making hematopoiesis a favorable system to study developmental cell lineage commitment. The main factors incorporating environmental signals to developing hematopoietic cells are cytokines, which regulate commitment of hematopoietic progenitors to the different blood lineages by acting either in an instructive or a permissive manner. Fms-like tyrosine kinase-3 (Flt3) ligand (FL) and Interleukin-7 (IL-7) are cytokines pivotal for B-cell development, as manifested by the severely compromised B-cell development in their absence. However, their precise role in regulating B-cell commitment has been the subject of debate. In the present study we assessed the rescue of B-cell commitment in mice lacking IL-7 but simultaneously overexpressing FL. Results obtained demonstrate that FL overexpression in IL-7-deficient mice rescues B-cell commitment, resulting in significant Ebf1 and Pax5 expression in Ly6D+CD135+CD127+CD19- precursors and subsequent generation of normal numbers of CD19+ B-cell progenitors, therefore indicating that IL-7 can be dispensable for commitment to the B-cell lineage. Further analysis of Ly6D+CD135+CD127+CD19- progenitors in IL-7- or FL-deficient mice overexpressing Bcl2, as well as in IL-7 transgenic mice suggests that both FL and IL-7 regulate B-cell commitment in a permissive manner: FL by inducing proliferation of Ly6D+CD135+CD127+CD19- progenitors and IL-7 by providing survival signals to these progenitors.
Keywords: commitment; cytokines; hematopoiesis; immunology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures














References
-
- Endele M, Etzrodt M, Schroeder T. Instruction of hematopoietic lineage choice by cytokine signaling. Exp Cell Res. 2014;329(2):207–213. - PubMed
-
- Enver T, Heyworth CM, Dexter TM. Do stem cells play dice? Blood. 1998;92(2):348–351, discussion 352. - PubMed
-
- Metcalf D. Lineage commitment and maturation in hematopoietic cells: The case for extrinsic regulation. Blood. 1998;92(2):345–347, discussion 352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

