Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder
- PMID: 27912058
- PMCID: PMC5554935
- DOI: 10.1016/j.cell.2016.11.013
Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder
Abstract
Autism spectrum disorders (ASD) are a group of genetic disorders often overlapping with other neurological conditions. We previously described abnormalities in the branched-chain amino acid (BCAA) catabolic pathway as a cause of ASD. Here, we show that the solute carrier transporter 7a5 (SLC7A5), a large neutral amino acid transporter localized at the blood brain barrier (BBB), has an essential role in maintaining normal levels of brain BCAAs. In mice, deletion of Slc7a5 from the endothelial cells of the BBB leads to atypical brain amino acid profile, abnormal mRNA translation, and severe neurological abnormalities. Furthermore, we identified several patients with autistic traits and motor delay carrying deleterious homozygous mutations in the SLC7A5 gene. Finally, we demonstrate that BCAA intracerebroventricular administration ameliorates abnormal behaviors in adult mutant mice. Our data elucidate a neurological syndrome defined by SLC7A5 mutations and support an essential role for the BCAA in human brain function.
Keywords: ASD; amino acid transporter; autism; blood brain barrier; excitation and inhibition imbalance; motor deficits.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


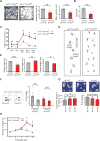


 ) or SLC7A5-A246G (●) and stopped at the indicated times (means ± SD; n=5 experiments). (D) Time dependence of [3H]His efflux from proteoliposomes reconstituted with SLC7A5-WT or SLC7A5-P375L was measured. Proteoliposomes reconstituted with SLC7A5-WT (○, ●) or SLC7A5-P375L (
) or SLC7A5-A246G (●) and stopped at the indicated times (means ± SD; n=5 experiments). (D) Time dependence of [3H]His efflux from proteoliposomes reconstituted with SLC7A5-WT or SLC7A5-P375L was measured. Proteoliposomes reconstituted with SLC7A5-WT (○, ●) or SLC7A5-P375L (
 ,
,
 ), containing 2 mM His were radioactivity-preloaded. Efflux was measured in absence (○,
), containing 2 mM His were radioactivity-preloaded. Efflux was measured in absence (○,
 ; uniport) or presence of external 1 mM His (●,
; uniport) or presence of external 1 mM His (●,
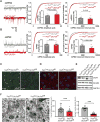
 ; antiport). Percentage of His efflux was calculated with respect to time 0 (means ± SD; n=4 experiments). (E) Radio-labeled leucine ([3H]Leu) transport analysis in human fibroblasts from affected (A) and unaffected (U) members of families 1426 and 1465 illustrating a significant reduction in leucine uptake by the cells of affected patients. Specificity of leucine uptake was assessed by competition with 10mM cold leucine (Leu) or 10 mM 2 BCH; ***P<0.001 (means ± SEM; N≥3 experiments performed with fibroblasts from two patients and one healthy control/family). See also Figure S7.
; antiport). Percentage of His efflux was calculated with respect to time 0 (means ± SD; n=4 experiments). (E) Radio-labeled leucine ([3H]Leu) transport analysis in human fibroblasts from affected (A) and unaffected (U) members of families 1426 and 1465 illustrating a significant reduction in leucine uptake by the cells of affected patients. Specificity of leucine uptake was assessed by competition with 10mM cold leucine (Leu) or 10 mM 2 BCH; ***P<0.001 (means ± SEM; N≥3 experiments performed with fibroblasts from two patients and one healthy control/family). See also Figure S7.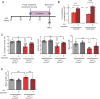
References
-
- Angelastro JM, Ignatova TN, Kukekov VG, Steindler DA, Stengren GB, Mendelsohn C, Greene LA. Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:4590–4600. - PMC - PubMed
-
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. - PubMed
-
- Banko JL, Klann E. Cap-dependent translation initiation and memory. Progress in brain research. 2008;169:59–80. - PubMed
-
- Braat S, Kooy RF. The GABAA Receptor as a Therapeutic Target for Neurodevelopmental Disorders. Neuron. 2015;86:1119–1130. - PubMed
-
- Braun LD, Cornford EM, Oldendorf WH. Newborn rabbit blood-brain barrier is selectively permeable and differs substantially from the adult. Journal of neurochemistry. 1980;34:147–152. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

