Modular Assembly of the Bacterial Large Ribosomal Subunit
- PMID: 27912064
- PMCID: PMC5145266
- DOI: 10.1016/j.cell.2016.11.020
Modular Assembly of the Bacterial Large Ribosomal Subunit
Abstract
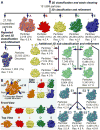
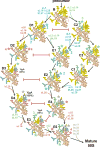
The ribosome is a complex macromolecular machine and serves as an ideal system for understanding biological macromolecular assembly. Direct observation of ribosome assembly in vivo is difficult, as few intermediates have been isolated and thoroughly characterized. Herein, we deploy a genetic system to starve cells of an essential ribosomal protein, which results in the accumulation of assembly intermediates that are competent for maturation. Quantitative mass spectrometry and single-particle cryo-electron microscopy reveal 13 distinct intermediates, which were each resolved to ∼4-5 Å resolution and could be placed in an assembly pathway. We find that ribosome biogenesis is a parallel process, that blocks of structured rRNA and proteins assemble cooperatively, and that the entire process is dynamic and can be "re-routed" through different pathways as needed. This work reveals the complex landscape of ribosome assembly in vivo and provides the requisite tools to characterize additional assembly pathways for ribosomes and other macromolecular machines.
Keywords: 50S subunit; RNA folding; Ribosome assembly; macromolecular assembly; quantitative mass spectrometry; single-particle cryo-electron microscopy.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
-
- Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457:977–980. - PubMed
-
- Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

