A Proteomic Approach to Analyze the Aspirin-mediated Lysine Acetylome
- PMID: 27913581
- PMCID: PMC5294217
- DOI: 10.1074/mcp.O116.065219
A Proteomic Approach to Analyze the Aspirin-mediated Lysine Acetylome
Abstract
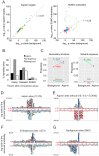
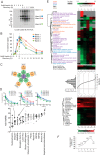
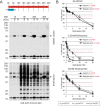
Aspirin, or acetylsalicylic acid is widely used to control pain, inflammation and fever. Important to this function is its ability to irreversibly acetylate cyclooxygenases at active site serines. Aspirin has the potential to acetylate other amino acid side-chains, leading to the possibility that aspirin-mediated lysine acetylation could explain some of its as-yet unexplained drug actions or side-effects. Using isotopically labeled aspirin-d3, in combination with acetylated lysine purification and LC-MS/MS, we identified over 12000 sites of lysine acetylation from cultured human cells. Although aspirin amplifies endogenous acetylation signals at the majority of detectable endogenous sites, cells tolerate aspirin mediated acetylation very well unless cellular deacetylases are inhibited. Although most endogenous acetylations are amplified by orders of magnitude, lysine acetylation site occupancies remain very low even after high doses of aspirin. This work shows that while aspirin has enormous potential to alter protein function, in the majority of cases aspirin-mediated acetylations do not accumulate to levels likely to elicit biological effects. These findings are consistent with an emerging model for cellular acetylation whereby stoichiometry correlates with biological relevance, and deacetylases act to minimize the biological consequences of nonspecific chemical acetylations.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Vane J. R., and Botting R. M. (2003) The mechanism of action of aspirin. Thromb. Res. 110, 255–258 - PubMed
-
- Brayfield A., ed. (2014) Martindale: The Complete Drug Reference., 38 ed., Pharmaceutical Press
-
- Algra A. M., and Rothwell P. M. (2012) Effects of regular aspirin on long-term cancer incidence and metastasis: a systematic comparison of evidence from observational studies versus randomised trials. Lancet Oncol. 13, 518–527 - PubMed
-
- Rothwell P. M., Price J. F., Fowkes F. G., Zanchetti A., Roncaglioni M. C., Tognoni G., Lee R., Belch J. F., Wilson M., Mehta Z., and Meade T. W. (2012) Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet 379, 1602–1612 - PubMed
-
- Rothwell P. M., Wilson M., Price J. F., Belch J. F., Meade T. W., and Mehta Z. (2012) Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet 379, 1591–1601 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

