Gene Regulation and Speciation
- PMID: 27914620
- PMCID: PMC5182078
- DOI: 10.1016/j.tig.2016.11.003
Gene Regulation and Speciation
Abstract
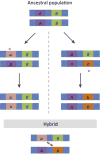
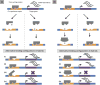
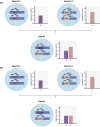
Understanding the genetic architecture of speciation is a major goal in evolutionary biology. Hybrid dysfunction is thought to arise most commonly through negative interactions between alleles at two or more loci. Divergence between interacting regulatory elements that affect gene expression (i.e., regulatory divergence) may be a common route for these negative interactions to arise. We review here how regulatory divergence between species can result in hybrid dysfunction, including recent theoretical support for this model. We then discuss the empirical evidence for regulatory divergence between species and evaluate evidence for misregulation as a source of hybrid dysfunction. Finally, we review unresolved questions in gene regulation as it pertains to speciation and point to areas that could benefit from future research.
Keywords: gene expression; gene regulation; hybrid inviability; hybrid sterility; speciation.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
References
-
- Lande R. Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution. 1979;33:234–251. - PubMed
-
- Hedrick PW. The establishment of chromosomal variants. Evolution. 1981;35:322–332. - PubMed
-
- Walsh JB. Rate of accumulation of reproductive isolation by chromosome rearrangements. Am Nat. 1982;120:510–532.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

