DNA damage related crosstalk between the nucleus and mitochondria
- PMID: 27915046
- PMCID: PMC5449269
- DOI: 10.1016/j.freeradbiomed.2016.11.050
DNA damage related crosstalk between the nucleus and mitochondria
Abstract
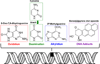
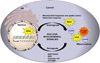
The electron transport chain is the primary pathway by which a cell generates energy in the form of ATP. Byproducts of this process produce reactive oxygen species that can cause damage to mitochondrial DNA. If not properly repaired, the accumulation of DNA damage can lead to mitochondrial dysfunction linked to several human disorders including neurodegenerative diseases and cancer. Mitochondria are able to combat oxidative DNA damage via repair mechanisms that are analogous to those found in the nucleus. Of the repair pathways currently reported in the mitochondria, the base excision repair pathway is the most comprehensively described. Proteins that are involved with the maintenance of mtDNA are encoded by nuclear genes and translocate to the mitochondria making signaling between the nucleus and mitochondria imperative. In this review, we discuss the current understanding of mitochondrial DNA repair mechanisms and also highlight the sensors and signaling pathways that mediate crosstalk between the nucleus and mitochondria in the event of mitochondrial stress.
Keywords: Mito-nuclear signaling; Mitochondrial DNA damage; Mitochondrial DNA repair; Mitochondrial dysfunction; Oxidative phosphorylation; REDOX signaling; Reactive oxygen species.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
None
Figures


References
-
- Kowaltowski AJ. Alternative mitochondrial functions in cell physiopathology: beyond ATP production. Braz J Med Biol Res. 2000;33(2):241–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

