Trends in Next-Generation Sequencing and a New Era for Whole Genome Sequencing
- PMID: 27915479
- PMCID: PMC5169091
- DOI: 10.5213/inj.1632742.371
Trends in Next-Generation Sequencing and a New Era for Whole Genome Sequencing
Abstract
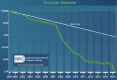
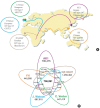
This article is a mini-review that provides a general overview for next-generation sequencing (NGS) and introduces one of the most popular NGS applications, whole genome sequencing (WGS), developed from the expansion of human genomics. NGS technology has brought massively high throughput sequencing data to bear on research questions, enabling a new era of genomic research. Development of bioinformatic software for NGS has provided more opportunities for researchers to use various applications in genomic fields. De novo genome assembly and large scale DNA resequencing to understand genomic variations are popular genomic research tools for processing a tremendous amount of data at low cost. Studies on transcriptomes are now available, from previous-hybridization based microarray methods. Epigenetic studies are also available with NGS applications such as whole genome methylation sequencing and chromatin immunoprecipitation followed by sequencing. Human genetics has faced a new paradigm of research and medical genomics by sequencing technologies since the Human Genome Project. The trend of NGS technologies in human genomics has brought a new era of WGS by enabling the building of human genomes databases and providing appropriate human reference genomes, which is a necessary component of personalized medicine and precision medicine.
Keywords: Epigenomics; Genomics; High-Throughput Nucleotide Sequencing; Human Genome Project; Sequence Analysis, RNA.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

