Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia
- PMID: 27916615
- PMCID: PMC5156313
- DOI: 10.1016/j.ccell.2016.11.002
Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia
Abstract
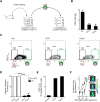
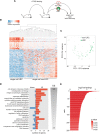
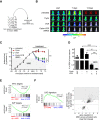
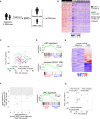
Tumor relapse is associated with dismal prognosis, but responsible biological principles remain incompletely understood. To isolate and characterize relapse-inducing cells, we used genetic engineering and proliferation-sensitive dyes in patient-derived xenografts of acute lymphoblastic leukemia (ALL). We identified a rare subpopulation that resembled relapse-inducing cells with combined properties of long-term dormancy, treatment resistance, and stemness. Single-cell and bulk expression profiling revealed their similarity to primary ALL cells isolated from pediatric and adult patients at minimal residual disease (MRD). Therapeutically adverse characteristics were reversible, as resistant, dormant cells became sensitive to treatment and started proliferating when dissociated from the in vivo environment. Our data suggest that ALL patients might profit from therapeutic strategies that release MRD cells from the niche.
Keywords: Cancer stem cells; RNA single-cell sequencing; acute lymphoblastic leukemia; dormant tumor cells; minimal residual disease (MRD); patient-derived xenograft (PDX) cells; primary patients' ALL MRD cells; treatment resistance.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Comment in
-
Leukaemia: Don't let sleeping cells lie.Nat Rev Cancer. 2016 Dec 21;17(1):3. doi: 10.1038/nrc.2016.150. Nat Rev Cancer. 2016. PMID: 27999430 No abstract available.
References
-
- Bacher U., Kohlmann A., Haferlach T. Gene expression profiling for diagnosis and therapy in acute leukaemia and other haematologic malignancies. Cancer Treat Rev. 2010;36:637–646. - PubMed
-
- Blatter S., Rottenberg S. Minimal residual disease in cancer therapy – small things make all the difference. Drug Resist Updat. 2015;21-22:1–10. - PubMed
-
- Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. - PubMed
-
- Castro Alves C., Terziyska N., Grunert M., Gündisch S., Graubner U., Quintanilla-Martinez L., Jeremias I. Leukemia-initiating cells of patient-derived acute lymphoblastic leukemia xenografts are sensitive toward TRAIL. Blood. 2012;119:4224–4227. - PubMed
-
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat. Med. 2011;17:313–319. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

