Pseudogenes and Their Genome-Wide Prediction in Plants
- PMID: 27916797
- PMCID: PMC5187791
- DOI: 10.3390/ijms17121991
Pseudogenes and Their Genome-Wide Prediction in Plants
Abstract
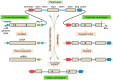
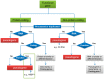
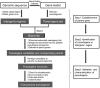
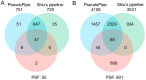
Pseudogenes are paralogs generated from ancestral functional genes (parents) during genome evolution, which contain critical defects in their sequences, such as lacking a promoter, having a premature stop codon or frameshift mutations. Generally, pseudogenes are functionless, but recent evidence demonstrates that some of them have potential roles in regulation. The majority of pseudogenes are generated from functional progenitor genes either by gene duplication (duplicated pseudogenes) or retro-transposition (processed pseudogenes). Pseudogenes are primarily identified by comparison to their parent genes. Bioinformatics tools for pseudogene prediction have been developed, among which PseudoPipe, PSF and Shiu's pipeline are publicly available. We compared these three tools using the well-annotated Arabidopsis thaliana genome and its known 924 pseudogenes as a test data set. PseudoPipe and Shiu's pipeline identified ~80% of A. thaliana pseudogenes, of which 94% were shared, while PSF failed to generate adequate results. A need for improvement of the bioinformatics tools for pseudogene prediction accuracy in plant genomes was thus identified, with the ultimate goal of improving the quality of genome annotation in plants.
Keywords: bioinformatics tools; duplicated; genome-wide; plants; processed; pseudogenes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

