Expression, Purification, and Characterization of Interleukin-11 Orthologues
- PMID: 27916836
- PMCID: PMC6274577
- DOI: 10.3390/molecules21121632
Expression, Purification, and Characterization of Interleukin-11 Orthologues
Abstract
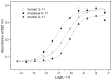
Interleukin-11 (IL-11) is a multifunctional cytokine implicated in several normal and pathological processes. The decoding of IL-11 function and development of IL-11-targeted drugs dictate the use of laboratory animals and need of the better understanding of species specificity of IL-11 signaling. Here, we present a method for the recombinant interleukin-11 (rIL-11) production from the important model animals, mouse and macaque. The purified mouse and macaque rIL-11 interact with extracellular domain of human IL-11 receptor subunit α and activate STAT3 signaling in HEK293 cells co-expressing human IL-11 receptors with efficacies resembling those of human rIL-11. Hence, the evolutionary divergence does not impair IL-11 signaling. Furthermore, compared to human rIL-11 its macaque orthologue is 8-fold more effective STAT3 activator, which favors its use for treatment of thrombocytopenia as a potent substitute for human rIL-11. Compared to IL-6, IL-11 signaling exhibits lower species specificity, likely due to less conserved intrinsic disorder propensity within IL-6 orthologues. The developed express method for preparation of functionally active macaque/mouse rIL-11 samples is suited for exploration of the molecular mechanisms underlying IL-11 action and for development of the drug candidates for therapy of oncologic/hematologic/inflammatory diseases related to IL-11 signaling.
Keywords: STAT3 signaling; bacterial expression; cloning; cytokines; interleukin-11; ligand-receptor interaction; protein-protein interaction; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Paul S.R., Bennett F., Calvetti J.A., Kelleher K., Wood C.R., O’Hara R.M., Jr., Leary A.C., Sibley B., Clark S.C., Williams D.A., et al. Molecular cloning of a cdna encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc. Natl. Acad. Sci. USA. 1990;87:7512–7516. doi: 10.1073/pnas.87.19.7512. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

