Characterization and 1.57 Å resolution structure of the key fire blight phosphatase AmsI from Erwinia amylovora
- PMID: 27917839
- PMCID: PMC5137468
- DOI: 10.1107/S2053230X16018781
Characterization and 1.57 Å resolution structure of the key fire blight phosphatase AmsI from Erwinia amylovora
Abstract
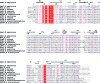
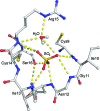
AmsI is a low-molecular-weight protein tyrosine phosphatase that regulates the production of amylovoran in the Gram-negative bacterium Erwinia amylovora, a specific pathogen of rosaceous plants such as apple, pear and quince. Amylovoran is an exopolysaccharide that is necessary for successful infection. In order to shed light on AmsI, its structure was solved at 1.57 Å resolution at the same pH as its highest measured activity (pH 5.5). In the active site, a water molecule, bridging between the catalytic Arg15 and the reaction-product analogue sulfate, might be representative of the water molecule attacking the phospho-cysteine intermediate in the second step of the reaction mechanism.
Keywords: Enterobacteriaceae; Erwinia amylovora; amylovoran; fire blight; hydrolases; kinetics; low-molecular-weight protein tyrosine phosphatase.
Figures

References
-
- Åkerud, T., Thulin, E., Van Etten, R. L. & Akke, M. (2002). J. Mol. Biol. 322, 137–152. - PubMed
-
- Bechet, E., Gruszczyk, J., Terreux, R., Gueguen-Chaignon, V., Vigouroux, A., Obadia, B., Cozzone, A. J., Nessler, S. & Grangeasse, C. (2010). Mol. Microbiol. 77, 1315–1325. - PubMed
-
- Bechet, E., Guiral, S., Torres, S., Mijakovic, I., Cozzone, A. J. & Grangeasse, C. (2009). Amino Acids, 37, 499–507. - PubMed
-
- Bellemann, P. & Geider, K. (1992). J. Gen. Microbiol. 138, 931–940. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

