Development of Cell-Permeable, Non-Helical Constrained Peptides to Target a Key Protein-Protein Interaction in Ovarian Cancer
- PMID: 27918136
- PMCID: PMC5291322
- DOI: 10.1002/anie.201609427
Development of Cell-Permeable, Non-Helical Constrained Peptides to Target a Key Protein-Protein Interaction in Ovarian Cancer
Abstract
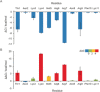
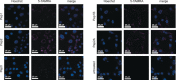
There is a lack of current treatment options for ovarian clear cell carcinoma (CCC) and the cancer is often resistant to platinum-based chemotherapy. Hence there is an urgent need for novel therapeutics. The transcription factor hepatocyte nuclear factor 1β (HNF1β) is ubiquitously overexpressed in CCC and is seen as an attractive therapeutic target. This was validated through shRNA-mediated knockdown of the target protein, HNF1β, in five high- and low-HNF1β-expressing CCC lines. To inhibit the protein function, cell-permeable, non-helical constrained proteomimetics to target the HNF1β-importin α protein-protein interaction were designed, guided by X-ray crystallographic data and molecular dynamics simulations. In this way, we developed the first reported series of constrained peptide nuclear import inhibitors. Importantly, this general approach may be extended to other transcription factors.
Keywords: constrained peptides; drug discovery; nuclear import; peptide therapeutics; peptidomimetics.
© 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures





References
-
- None
-
- None
-
- Kajihara H., Yamada Y., Kanayama S., Furukawa N., Noguchi T., Haruta S., Yoshida S., Sado T., Oi H., Kobayashi H., Oncol. Rep. 2010, 23, 1193–1203; - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

