A Review of Computational Methods for Finding Non-Coding RNA Genes
- PMID: 27918472
- PMCID: PMC5192489
- DOI: 10.3390/genes7120113
A Review of Computational Methods for Finding Non-Coding RNA Genes
Abstract
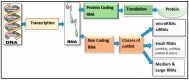
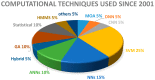
Finding non-coding RNA (ncRNA) genes has emerged over the past few years as a cutting-edge trend in bioinformatics. There are numerous computational intelligence (CI) challenges in the annotation and interpretation of ncRNAs because it requires a domain-related expert knowledge in CI techniques. Moreover, there are many classes predicted yet not experimentally verified by researchers. Recently, researchers have applied many CI methods to predict the classes of ncRNAs. However, the diverse CI approaches lack a definitive classification framework to take advantage of past studies. A few review papers have attempted to summarize CI approaches, but focused on the particular methodological viewpoints. Accordingly, in this article, we summarize in greater detail than previously available, the CI techniques for finding ncRNAs genes. We differentiate from the existing bodies of research and discuss concisely the technical merits of various techniques. Lastly, we review the limitations of ncRNA gene-finding CI methods with a point-of-view towards the development of new computational tools.
Keywords: Bayesian networks; DNA; computational intelligence; deep learning; gene; genetic algorithm; micro RNA; neural network; non-coding RNA; support vector machine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

