Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells
- PMID: 27918527
- PMCID: PMC5488270
- DOI: 10.1038/nmicrobiol.2016.250
Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells
Abstract
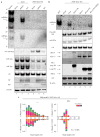
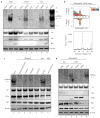
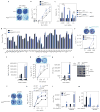
Influenza A virus (IAV) causes annual epidemics and occasional pandemics, and is one of the best-characterized human RNA viral pathogens1. However, a physiologically relevant role for the RNA interference (RNAi) suppressor activity of the IAV non-structural protein 1 (NS1), reported over a decade ago2, remains unknown3. Plant and insect viruses have evolved diverse virulence proteins to suppress RNAi as their hosts produce virus-derived small interfering RNAs (siRNAs) that direct specific antiviral defence4-7 by an RNAi mechanism dependent on the slicing activity of Argonaute proteins (AGOs)8,9. Recent studies have documented induction and suppression of antiviral RNAi in mouse embryonic stem cells and suckling mice10,11. However, it is still under debate whether infection by IAV or any other RNA virus that infects humans induces and/or suppresses antiviral RNAi in mature mammalian somatic cells12-21. Here, we demonstrate that mature human somatic cells produce abundant virus-derived siRNAs co-immunoprecipitated with AGOs in response to IAV infection. We show that the biogenesis of viral siRNAs from IAV double-stranded RNA (dsRNA) precursors in infected cells is mediated by wild-type human Dicer and potently suppressed by both NS1 of IAV as well as virion protein 35 (VP35) of Ebola and Marburg filoviruses. We further demonstrate that the slicing catalytic activity of AGO2 inhibits IAV and other RNA viruses in mature mammalian cells, in an interferon-independent fashion. Altogether, our work shows that IAV infection induces and suppresses antiviral RNAi in differentiated mammalian somatic cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Questioning antiviral RNAi in mammals.Nat Microbiol. 2017 Apr 25;2:17052. doi: 10.1038/nmicrobiol.2017.52. Nat Microbiol. 2017. PMID: 28440277 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

