Laboratory Diagnosis of Chikungunya Virus Infections and Commercial Sources for Diagnostic Assays
- PMID: 27920176
- PMCID: PMC5657184
- DOI: 10.1093/infdis/jiw274
Laboratory Diagnosis of Chikungunya Virus Infections and Commercial Sources for Diagnostic Assays
Abstract
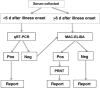
Detection of chikungunya virus (CHIKV) or viral RNA is the primary laboratory test used to diagnose infection in serum collected <6 days after onset of illness. Two real-time reverse transcription-polymerase chain reaction (RT-PCR) kits are available commercially, but validity data are limited. There are 2 commercial sources of inactivated positive-control CHIKV RNA to be used with purchased primers. The Centers for Disease Control and Prevention provides viral RNA-positive controls and primer and probe nucleotide sequences for real-time RT-PCR testing. Detection of CHIKV-specific immunoglobulin M (IgM) antibody becomes a sensitive test for samples collected approximately >5 days of illness. Commercially available CHIKV IgM-detection assays include lateral flow rapid tests, IgM antibody capture enzyme-linked immunosorbent assays (MAC-ELISAs), and indirect immunofluorescence tests. Nine commercial CHIKV IgM detection assays were evaluated at 3 reference laboratories to provide guidance to public health diagnostic laboratories on their performance parameters. Sensitivity of the rapid tests and 3 MAC-ELISAs was <50%, and thus these assays are not recommended. Three of the MAC-ELISA kits and 1 indirect immunofluorescence kit had comparable performance to the reference assays. In summary, commercial assays with performance comparable to reference assays are available for molecular and serological diagnosis of CHIKV infections.
Keywords: Chikungunya virus; IgM antibody capture enzyme-linked immunosorbent assay (MAC-ELISA); arbovirus diagnostic testing; real-time RT-PCR.
Published by Oxford University Press for the Infectious Diseases Society of America 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Conflict of interest statement
Figures

References
-
- Cassadou S, Boucau S, Petit-Sinturel M, Huc P, Leparc-Goffart I, Ledrans M. Emergence of chikungunya fever on the French side of Saint Martin island, October to December 2013. Euro Surveill. 2014;19:13–6. - PubMed
-
- Lepiniec L, Dalgarno L, Huong VT, Monath TP, Digoutte JP, Deubel V. Geographic distribution and evolution of yellow fever viruses based on direct sequencing of genomic cDNA fragments. J Gen Virol. 1994;75:417–23. - PubMed
-
- Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49:942–8. - PubMed
-
- Powers AM. Chikungunya. Clin in Lab Med. 2010;30:209–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

