The Streptococcus gordonii Adhesin CshA Protein Binds Host Fibronectin via a Catch-Clamp Mechanism
- PMID: 27920201
- PMCID: PMC5290933
- DOI: 10.1074/jbc.M116.760975
The Streptococcus gordonii Adhesin CshA Protein Binds Host Fibronectin via a Catch-Clamp Mechanism
Abstract
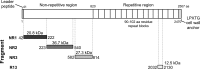
Adherence of bacteria to biotic or abiotic surfaces is a prerequisite for host colonization and represents an important step in microbial pathogenicity. This attachment is facilitated by bacterial adhesins at the cell surface. Because of their size and often elaborate multidomain architectures, these polypeptides represent challenging targets for detailed structural and functional characterization. The multifunctional fibrillar adhesin CshA, which mediates binding to both host molecules and other microorganisms, is an important determinant of colonization by Streptococcus gordonii, an oral commensal and opportunistic pathogen of animals and humans. CshA binds the high-molecular-weight glycoprotein fibronectin (Fn) via an N-terminal non-repetitive region, and this protein-protein interaction has been proposed to promote S. gordonii colonization at multiple sites within the host. However, the molecular details of how these two proteins interact have yet to be established. Here we present a structural description of the Fn binding N-terminal region of CshA, derived from a combination of X-ray crystallography, small angle X-ray scattering, and complementary biophysical methods. In vitro binding studies support a previously unreported two-state "catch-clamp" mechanism of Fn binding by CshA, in which the disordered N-terminal domain of CshA acts to "catch" Fn, via formation of a rapidly assembled but also readily dissociable pre-complex, enabling its neighboring ligand binding domain to tightly clamp the two polypeptides together. This study presents a new paradigm for target binding by a bacterial adhesin, the identification of which will inform future efforts toward the development of anti-adhesive agents that target S. gordonii and related streptococci.
Keywords: X-ray crystallography; adhesin; bacterial pathogenesis; intrinsically disordered protein; microbiology; small-angle X-ray scattering (SAXS).
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Comment in
-
Bacterium 'Lasso' From Mouth to Heart Can Cause Disease.J Calif Dent Assoc. 2017 Apr;45(4):175. J Calif Dent Assoc. 2017. PMID: 29068617 No abstract available.
Similar articles
-
Multiple adhesin proteins on the cell surface of Streptococcus gordonii are involved in adhesion to human fibronectin.Microbiology (Reading). 2009 Nov;155(Pt 11):3572-3580. doi: 10.1099/mic.0.032078-0. Epub 2009 Aug 6. Microbiology (Reading). 2009. PMID: 19661180 Free PMC article.
-
The streptococcal multidomain fibrillar adhesin CshA has an elongated polymeric architecture.J Biol Chem. 2020 May 8;295(19):6689-6699. doi: 10.1074/jbc.RA119.011719. Epub 2020 Mar 30. J Biol Chem. 2020. PMID: 32229583 Free PMC article.
-
Domain shuffling of a highly mutable ligand-binding fold drives adhesin generation across the bacterial kingdom.Proteins. 2023 Aug;91(8):1007-1020. doi: 10.1002/prot.26487. Epub 2023 Mar 20. Proteins. 2023. PMID: 36912614 Free PMC article.
-
[Adhesins of oral streptococci].Nihon Saikingaku Zasshi. 2013;68(2):283-93. doi: 10.3412/jsb.68.283. Nihon Saikingaku Zasshi. 2013. PMID: 23727707 Review. Japanese.
-
Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells.Matrix Biol. 1999 Jun;18(3):211-23. doi: 10.1016/s0945-053x(99)00025-6. Matrix Biol. 1999. PMID: 10429941 Review.
Cited by
-
The Group B Streptococcal surface antigen I/II protein, BspC, interacts with host vimentin to promote adherence to brain endothelium and inflammation during the pathogenesis of meningitis.PLoS Pathog. 2019 Jun 10;15(6):e1007848. doi: 10.1371/journal.ppat.1007848. eCollection 2019 Jun. PLoS Pathog. 2019. PMID: 31181121 Free PMC article.
-
Fibronectin and Its Role in Human Infective Diseases.Cells. 2019 Nov 26;8(12):1516. doi: 10.3390/cells8121516. Cells. 2019. PMID: 31779172 Free PMC article. Review.
-
Gymnemic Acids Inhibit Adhesive Nanofibrillar Mediated Streptococcus gordonii-Candida albicans Mono-Species and Dual-Species Biofilms.Front Microbiol. 2019 Oct 11;10:2328. doi: 10.3389/fmicb.2019.02328. eCollection 2019. Front Microbiol. 2019. PMID: 31681200 Free PMC article.
-
Novel Virulent Bacteriophage ΦSG005, Which Infects Streptococcus gordonii, Forms a Distinct Clade among Streptococcus Viruses.Viruses. 2021 Sep 29;13(10):1964. doi: 10.3390/v13101964. Viruses. 2021. PMID: 34696394 Free PMC article.
-
Streptococcus gordonii: Pathogenesis and Host Response to Its Cell Wall Components.Microorganisms. 2020 Nov 24;8(12):1852. doi: 10.3390/microorganisms8121852. Microorganisms. 2020. PMID: 33255499 Free PMC article. Review.
References
-
- Kline K. A., Fälker S., Dahlberg S., Normark S., and Henriques-Normark B. (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 - PubMed
-
- Demuyser L., Jabra-Rizk M. A., and Van Dijck P. (2014) Microbial cell surface proteins and secreted metabolites involved in multispecies biofilms. Pathog. Dis. 70, 219–230 - PubMed
-
- Löfling J., Vimberg V., Battig P., and Henriques-Normark B. (2011) Cellular interactions by LPxTG-anchored pneumococcal adhesins and their streptococcal homologues. Cell. Microbiol. 13, 186–197 - PubMed
-
- Larson M. R., Rajashankar K. R., Patel M. H., Robinette R. A., Crowley P. J., Michalek S., Brady L. J., and Deivanayagam C. (2010) Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of α- and PPII-helices. Proc. Natl. Acad. Sci. U.S.A. 107, 5983–5988 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE016690/DE/NIDCR NIH HHS/United States
- PG/17/10/32829/BHF_/British Heart Foundation/United Kingdom
- R01 DE012505/DE/NIDCR NIH HHS/United States
- BB/I006478/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L01386X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

