A Molecular Basis for the Presentation of Phosphorylated Peptides by HLA-B Antigens
- PMID: 27920218
- PMCID: PMC5294207
- DOI: 10.1074/mcp.M116.063800
A Molecular Basis for the Presentation of Phosphorylated Peptides by HLA-B Antigens
Abstract
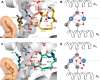
As aberrant protein phosphorylation is a hallmark of tumor cells, the display of tumor-specific phosphopeptides by Human Leukocyte Antigen (HLA) class I molecules can be exploited in the treatment of cancer by T-cell-based immunotherapy. Yet, the characterization and prediction of HLA-I phospholigands is challenging as the molecular determinants of the presentation of such post-translationally modified peptides are not fully understood. Here, we employed a peptidomic workflow to identify 256 unique phosphorylated ligands associated with HLA-B*40, -B*27, -B*39, or -B*07. Remarkably, these phosphopeptides showed similar molecular features. Besides the specific anchor motifs imposed by the binding groove of each allotype, the predominance of phosphorylation at peptide position 4 (P4) became strikingly evident, as was the enrichment of basic residues at P1. To determine the structural basis of this observation, we carried out a series of peptide binding assays and solved the crystal structures of HLA-B*40 in complex with a phosphorylated ligand or its nonphosphorylated counterpart. Overall, our data provide a clear explanation to the common motif found in the phosphopeptidomes associated to different HLA-B molecules. The high prevalence of phosphorylation at P4 is dictated by the presence of the conserved residue Arg62 in the heavy chain, a structural feature shared by most HLA-B alleles. In contrast, the preference for basic residues at P1 is allotype-dependent and might be linked to the structure of the A pocket. This molecular understanding of the presentation of phosphopeptides by HLA-B molecules provides a base for the improved prediction and identification of phosphorylated neo-antigens, as potentially used for cancer immunotherapy.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

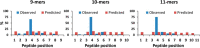



Similar articles
-
The landscape of MHC-presented phosphopeptides yields actionable shared tumor antigens for cancer immunotherapy across multiple HLA alleles.bioRxiv [Preprint]. 2023 Feb 12:2023.02.08.527552. doi: 10.1101/2023.02.08.527552. bioRxiv. 2023. Update in: J Immunother Cancer. 2023 Sep;11(9):e006889. doi: 10.1136/jitc-2023-006889. PMID: 36798179 Free PMC article. Updated. Preprint.
-
Increased diversity of the HLA-B40 ligandome by the presentation of peptides phosphorylated at their main anchor residue.Mol Cell Proteomics. 2014 Feb;13(2):462-74. doi: 10.1074/mcp.M113.034314. Epub 2013 Dec 23. Mol Cell Proteomics. 2014. PMID: 24366607 Free PMC article.
-
Position 45 influences the peptide binding motif of HLA-B*44:08.Immunogenetics. 2012 Mar;64(3):245-9. doi: 10.1007/s00251-011-0583-z. Epub 2011 Oct 19. Immunogenetics. 2012. PMID: 22009320
-
Naturally occurring A pocket polymorphism in HLA-B*2703 increases the dependence on an accessory anchor residue at P1 for optimal binding of nonamer peptides.J Immunol. 1997 Nov 15;159(10):4887-97. J Immunol. 1997. PMID: 9366414
-
Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype.Acta Crystallogr D Biol Crystallogr. 2006 Nov;62(Pt 11):1300-10. doi: 10.1107/S0907444906027636. Epub 2006 Oct 18. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 17057332
Cited by
-
AlphaPeptDeep: a modular deep learning framework to predict peptide properties for proteomics.Nat Commun. 2022 Nov 24;13(1):7238. doi: 10.1038/s41467-022-34904-3. Nat Commun. 2022. PMID: 36433986 Free PMC article.
-
The landscape of MHC-presented phosphopeptides yields actionable shared tumor antigens for cancer immunotherapy across multiple HLA alleles.bioRxiv [Preprint]. 2023 Feb 12:2023.02.08.527552. doi: 10.1101/2023.02.08.527552. bioRxiv. 2023. Update in: J Immunother Cancer. 2023 Sep;11(9):e006889. doi: 10.1136/jitc-2023-006889. PMID: 36798179 Free PMC article. Updated. Preprint.
-
Strategies for Non-Covalent Attachment of Antibodies to PEGylated Nanoparticles for Targeted Drug Delivery.Int J Nanomedicine. 2024 Oct 1;19:10045-10064. doi: 10.2147/IJN.S479270. eCollection 2024. Int J Nanomedicine. 2024. PMID: 39371476 Free PMC article. Review.
-
Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry.Immunology. 2018 Jul;154(3):331-345. doi: 10.1111/imm.12936. Epub 2018 May 8. Immunology. 2018. PMID: 29658117 Free PMC article. Review.
-
Molecular mechanism of phosphopeptide neoantigen immunogenicity.Nat Commun. 2023 Jun 23;14(1):3763. doi: 10.1038/s41467-023-39425-1. Nat Commun. 2023. PMID: 37353482 Free PMC article.
References
-
- Robinson J., Soormally A. R., Hayhurst J. D., and Marsh S. G. (2016) The IPD-IMGT/HLA Database - New developments in reporting HLA variation. Human Immunol. 77, 233–237 - PubMed
-
- Engelhard V. H., Altrich-Vanlith M., Ostankovitch M., and Zarling A. L. (2006) Post-translational modifications of naturally processed MHC-binding epitopes. Cur. Opin. Immunol. 18, 92–97 - PubMed
-
- Meyer V. S., Drews O., Gunder M., Hennenlotter J., Rammensee H. G., and Stevanovic S. (2009) Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J. Proteome Res. 8, 3666–3674 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

