One-Step PCR Detection of Salmonella Pullorum/Gallinarum Using a Novel Target: The Flagellar Biosynthesis Gene flhB
- PMID: 27920764
- PMCID: PMC5118417
- DOI: 10.3389/fmicb.2016.01863
One-Step PCR Detection of Salmonella Pullorum/Gallinarum Using a Novel Target: The Flagellar Biosynthesis Gene flhB
Abstract
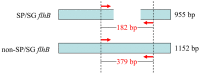
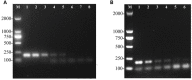
Salmonella enterica serovar Pullorum/Gallinarum is an important infectious pathogen that has caused widespread problems for chicken industry. Traditional Salmonella serotyping is an expensive and time-consuming process. In this study, we developed a rapid one-step polymerase chain reaction (PCR) method to identify S. Pullorum/Gallinarum. The PCR-based assay focuses on flhB, which shows a deficient region only in S. Pullorum/Gallinarum, compared with that of other serovars. The specificity and sensitivity of the PCR system were evaluated. The developed PCR method could identify S. Pullorum/Gallinarum from 27 different Salmonella serovars and eight non-Salmonella pathogens. The minimum limit of DNA and the lowest number of cells of S. Pullorum for the PCR detection were no less than 5.85 pg/μL and 10 CFU, respectively. The method was applied to the analysis of Salmonella strains isolated from the chicken farm. The PCR-based testing results of the farm isolates were in concordance with those obtained using traditional serotyping method. This newly developed PCR-based system could be used to accurately screen for the presence of S. Pullorum/Gallinarum, and support traditional serotyping methods, especially in high-throughput screening situations.
Keywords: PCR detection; Salmonella Pullorum/Gallinarum; chicken farm; flhB; one-step.
Figures


Similar articles
-
An Efficient Multiplex PCR-Based Assay as a Novel Tool for Accurate Inter-Serovar Discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S. Dublin.Front Microbiol. 2017 Mar 16;8:420. doi: 10.3389/fmicb.2017.00420. eCollection 2017. Front Microbiol. 2017. PMID: 28360901 Free PMC article.
-
Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum in Poultry: Review of Pathogenesis, Antibiotic Resistance, Diagnosis and Control in the Genomic Era.Antibiotics (Basel). 2023 Dec 25;13(1):23. doi: 10.3390/antibiotics13010023. Antibiotics (Basel). 2023. PMID: 38247582 Free PMC article. Review.
-
A new PCR assay based on the new gene-SPUL_2693 for rapid detection of Salmonella enterica subsp. enterica serovar Gallinarum biovars Gallinarum and Pullorum.Poult Sci. 2018 Nov 1;97(11):4000-4007. doi: 10.3382/ps/pey254. Poult Sci. 2018. PMID: 30101343
-
Identification and Discrimination of Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum Based on a One-Step Multiplex PCR Assay.Front Microbiol. 2018 Jul 31;9:1718. doi: 10.3389/fmicb.2018.01718. eCollection 2018. Front Microbiol. 2018. PMID: 30108571 Free PMC article.
-
The immunobiology of avian systemic salmonellosis.Vet Immunol Immunopathol. 2009 Mar 15;128(1-3):53-9. doi: 10.1016/j.vetimm.2008.10.295. Epub 2008 Oct 17. Vet Immunol Immunopathol. 2009. PMID: 19070366 Review.
Cited by
-
An Efficient Multiplex PCR-Based Assay as a Novel Tool for Accurate Inter-Serovar Discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S. Dublin.Front Microbiol. 2017 Mar 16;8:420. doi: 10.3389/fmicb.2017.00420. eCollection 2017. Front Microbiol. 2017. PMID: 28360901 Free PMC article.
-
CRISPR 2 PCR and high resolution melting profiling for identification and characterization of clinically-relevant Salmonella enterica subsp. enterica.PeerJ. 2020 Jun 16;8:e9113. doi: 10.7717/peerj.9113. eCollection 2020. PeerJ. 2020. PMID: 32587791 Free PMC article.
-
Fowl Typhoid Outbreak on a Commercial Turkey Farm in Croatia.Microorganisms. 2024 Jan 13;12(1):165. doi: 10.3390/microorganisms12010165. Microorganisms. 2024. PMID: 38257990 Free PMC article.
-
Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum in Poultry: Review of Pathogenesis, Antibiotic Resistance, Diagnosis and Control in the Genomic Era.Antibiotics (Basel). 2023 Dec 25;13(1):23. doi: 10.3390/antibiotics13010023. Antibiotics (Basel). 2023. PMID: 38247582 Free PMC article. Review.
-
The effect of oregano essential oil on the prevention and treatment of Salmonella pullorum and Salmonella gallinarum infections in commercial Yellow-chicken breeders.Front Vet Sci. 2022 Dec 21;9:1058844. doi: 10.3389/fvets.2022.1058844. eCollection 2022. Front Vet Sci. 2022. PMID: 36619954 Free PMC article.
References
-
- Grimont P. A. D., Weill F.-X. (2007). Antigenic Formulae of the Salmonella serovars 9th Edn. Paris: WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

