Transcriptional Profiling of Type II Toxin-Antitoxin Genes of Helicobacter pylori under Different Environmental Conditions: Identification of HP0967-HP0968 System
- PMID: 27920769
- PMCID: PMC5118875
- DOI: 10.3389/fmicb.2016.01872
Transcriptional Profiling of Type II Toxin-Antitoxin Genes of Helicobacter pylori under Different Environmental Conditions: Identification of HP0967-HP0968 System
Abstract
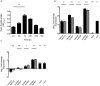
Helicobacter pylori is a Gram-negative bacterium that colonizes the human gastric mucosa and is responsible for causing peptic ulcers and gastric carcinoma. The expression of virulence factors allows the persistence of H. pylori in the stomach, which results in a chronic, sometimes uncontrolled inflammatory response. Type II toxin-antitoxin (TA) systems have emerged as important virulence factors in many pathogenic bacteria. Three type II TA systems have previously been identified in the genome of H. pylori 26695: HP0315-HP0316, HP0892-HP0893, and HP0894-HP0895. Here we characterized a heretofore undescribed type II TA system in H. pylori, HP0967-HP0968, which is encoded by the bicistronic operon hp0968-hp0967 and belongs to the Vap family. The predicted HP0967 protein is a toxin with ribonuclease activity whereas HP0968 is an antitoxin that binds to its own regulatory region. We found that all type II TA systems were expressed in H. pylori during early stationary growth phase, and differentially expressed in the presence of urea, nickel, and iron, although, the hp0968-hp0967 pair was the most affected under these environmental conditions. Transcription of hp0968-hp0967 was strongly induced in a mature H. pylori biofilm and when the bacteria interacted with AGS epithelial cells. Kanamycin and chloramphenicol considerably boosted transcription levels of all the four type II TA systems. The hp0968-hp0967 TA system was the most frequent among 317 H. pylori strains isolated from all over the world. This study is the first report on the transcription of type II TA genes in H. pylori under different environmental conditions. Our data show that the HP0967 and HP0968 proteins constitute a bona fide type II TA system in H. pylori, whose expression is regulated by environmental cues, which are relevant in the context of infection of the human gastric mucosa.
Keywords: H. pylori; HP0967; HP0968; environmental cues; toxin–antitoxin system.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

