Genome-Wide Association Study for Identifying Loci that Affect Fillet Yield, Carcass, and Body Weight Traits in Rainbow Trout (Oncorhynchus mykiss)
- PMID: 27920797
- PMCID: PMC5118429
- DOI: 10.3389/fgene.2016.00203
Genome-Wide Association Study for Identifying Loci that Affect Fillet Yield, Carcass, and Body Weight Traits in Rainbow Trout (Oncorhynchus mykiss)
Abstract
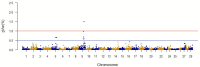
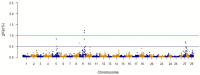
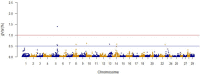
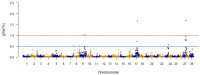
Fillet yield (FY, %) is an economically-important trait in rainbow trout aquaculture that affects production efficiency. Despite that, FY has received little attention in breeding programs because it is difficult to measure on a large number of fish and cannot be directly measured on breeding candidates. The recent development of a high-density SNP array for rainbow trout has provided the needed tool for studying the underlying genetic architecture of this trait. A genome-wide association study (GWAS) was conducted for FY, body weight at 10 (BW10) and 13 (BW13) months post-hatching, head-off carcass weight (CAR), and fillet weight (FW) in a pedigreed rainbow trout population selectively bred for improved growth performance. The GWAS analysis was performed using the weighted single-step GBLUP method (wssGWAS). Phenotypic records of 1447 fish (1.5 kg at harvest) from 299 full-sib families in three successive generations, of which 875 fish from 196 full-sib families were genotyped, were used in the GWAS analysis. A total of 38,107 polymorphic SNPs were analyzed in a univariate model with hatch year and harvest group as fixed effects, harvest weight as a continuous covariate, and animal and common environment as random effects. A new linkage map was developed to create windows of 20 adjacent SNPs for use in the GWAS. The two windows with largest effect for FY and FW were located on chromosome Omy9 and explained only 1.0-1.5% of genetic variance, thus suggesting a polygenic architecture affected by multiple loci with small effects in this population. One window on Omy5 explained 1.4 and 1.0% of the genetic variance for BW10 and BW13, respectively. Three windows located on Omy27, Omy17, and Omy9 (same window detected for FY) explained 1.7, 1.7, and 1.0%, respectively, of genetic variance for CAR. Among the detected 100 SNPs, 55% were located directly in genes (intron and exons). Nucleotide sequences of intragenic SNPs were blasted to the Mus musculus genome to create a putative gene network. The network suggests that differences in the ability to maintain a proliferative and renewable population of myogenic precursor cells may affect variation in growth and fillet yield in rainbow trout.
Keywords: fillet yield; gene network; genome-wide association study; genomic selection; linkage map; rainbow trout; single-step GBLUP.
Figures


Similar articles
-
Genomic selection models substantially improve the accuracy of genetic merit predictions for fillet yield and body weight in rainbow trout using a multi-trait model and multi-generation progeny testing.Genet Sel Evol. 2023 Feb 9;55(1):11. doi: 10.1186/s12711-023-00782-6. Genet Sel Evol. 2023. PMID: 36759760 Free PMC article.
-
Genome-wide association analysis for body weight identifies candidate genes related to development and metabolism in rainbow trout (Oncorhynchus mykiss).Mol Genet Genomics. 2019 Jun;294(3):563-571. doi: 10.1007/s00438-018-1518-2. Epub 2019 Jan 11. Mol Genet Genomics. 2019. PMID: 30635785
-
Genome-wide identification of loci associated with growth in rainbow trout.BMC Genomics. 2020 Mar 5;21(1):209. doi: 10.1186/s12864-020-6617-x. BMC Genomics. 2020. PMID: 32138655 Free PMC article.
-
Weighted Single-Step GWAS Identifies Genes Influencing Fillet Color in Rainbow Trout.Genes (Basel). 2022 Jul 26;13(8):1331. doi: 10.3390/genes13081331. Genes (Basel). 2022. PMID: 35893068 Free PMC article.
-
First Evidence of Realized Selection Response on Fillet Yield in Rainbow Trout Oncorhynchus mykiss, Using Sib Selection or Based on Correlated Ultrasound Measurements.Front Genet. 2019 Dec 20;10:1225. doi: 10.3389/fgene.2019.01225. eCollection 2019. Front Genet. 2019. PMID: 31921286 Free PMC article.
Cited by
-
First genomic prediction and genome-wide association for complex growth-related traits in Rock Bream (Oplegnathus fasciatus).Evol Appl. 2021 Mar 17;15(4):523-536. doi: 10.1111/eva.13218. eCollection 2022 Apr. Evol Appl. 2021. PMID: 35505886 Free PMC article.
-
Development of genomic predictions for harvest and carcass weight in channel catfish.Genet Sel Evol. 2018 Dec 14;50(1):66. doi: 10.1186/s12711-018-0435-5. Genet Sel Evol. 2018. PMID: 30547740 Free PMC article.
-
Genome Wide Analysis for Growth at Two Growth Stages in A New Fast-Growing Common Carp Strain (Cyprinus carpio L.).Sci Rep. 2020 Apr 29;10(1):7259. doi: 10.1038/s41598-020-64037-w. Sci Rep. 2020. PMID: 32350307 Free PMC article.
-
Identification of SNPs associated with muscle yield and quality traits using allelic-imbalance analyses of pooled RNA-Seq samples in rainbow trout.BMC Genomics. 2017 Aug 7;18(1):582. doi: 10.1186/s12864-017-3992-z. BMC Genomics. 2017. PMID: 28784089 Free PMC article.
-
Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research.BMC Genomics. 2017 Feb 20;18(1):191. doi: 10.1186/s12864-017-3557-1. BMC Genomics. 2017. PMID: 28219347 Free PMC article. Review.
References
-
- Aguilar I., Misztal I., Tsuruta S., Legarra A., Wang H. (2014). PREGSF90 – POSTGSF90: computational tools for the implementation of single-step genomic selection and genome-wide association with ungenotyped individuals in BLUPF90 programs, in Proceedings, 10th World Congress of Genetics Applied to Livestock Production (Vancouver, BC: ).
-
- Atencio I. A., Ramachandra M., Shabram P., Demers G. W. (2000). Calpain inhibitor 1 activates p53-dependent apoptosis in tumor cell lines. Cell Growth Differ. 11, 247–253. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

