Activation of dioxygen by copper metalloproteins and insights from model complexes
- PMID: 27921179
- PMCID: PMC5600896
- DOI: 10.1007/s00775-016-1415-2
Activation of dioxygen by copper metalloproteins and insights from model complexes
Abstract
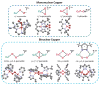
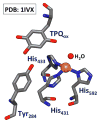
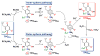
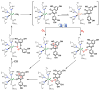
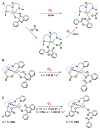
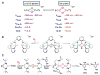
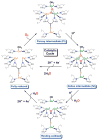
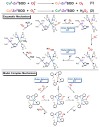
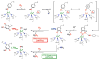
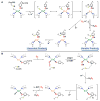
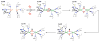
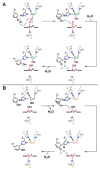
Nature uses dioxygen as a key oxidant in the transformation of biomolecules. Among the enzymes that are utilized for these reactions are copper-containing metalloenzymes, which are responsible for important biological functions such as the regulation of neurotransmitters, dioxygen transport, and cellular respiration. Enzymatic and model system studies work in tandem in order to gain an understanding of the fundamental reductive activation of dioxygen by copper complexes. This review covers the most recent advancements in the structures, spectroscopy, and reaction mechanisms for dioxygen-activating copper proteins and relevant synthetic models thereof. An emphasis has also been placed on cofactor biogenesis, a fundamentally important process whereby biomolecules are post-translationally modified by the pro-enzyme active site to generate cofactors which are essential for the catalytic enzymatic reaction. Significant questions remaining in copper-ion-mediated O2-activation in copper proteins are addressed.
Keywords: Cofactor biogenesis; Copper; Copper enzymes; Dioxygen activation; Enzymatic mechanisms.
Figures
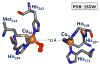


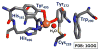
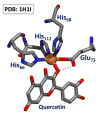


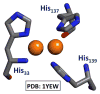




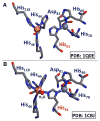













References
-
- Kroneck PMH, Torres MES, editors. Sustaining life on planet earth: metalloenzymes mastering dioxygen and other chewy gases. Springer International Publishing; Switzerland: 2015. - PubMed
-
- Mccann SD, Stahl SS. Acc Chem Res. 2015;48:1756–1766. - PubMed
-
- Gubelmann MH, Williams AF. Struct Bonding. Vol. 55. Berlin: 1983. pp. 1–65.
-
- Sawyer DT. Oxygen chemistry. Oxford University Press; New York: 1991.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

