MALAT1 and HOTAIR Long Non-Coding RNAs Play Opposite Role in Estrogen-Mediated Transcriptional Regulation in Prostate Cancer Cells
- PMID: 27922078
- PMCID: PMC5138831
- DOI: 10.1038/srep38414
MALAT1 and HOTAIR Long Non-Coding RNAs Play Opposite Role in Estrogen-Mediated Transcriptional Regulation in Prostate Cancer Cells
Abstract
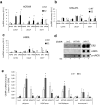
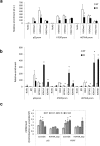
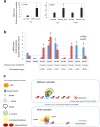
In the complex network of nuclear hormone receptors, the long non-coding RNAs (lncRNAs) are emerging as critical determinants of hormone action. Here we investigated the involvement of selected cancer-associated lncRNAs in Estrogen Receptor (ER) signaling. Prior studies by Chromatin Immunoprecipitation (ChIP) Sequencing showed that in prostate cancer cells ERs form a complex with the endothelial nitric oxide synthase (eNOS) and that in turn these complexes associate with chromatin in an estrogen-dependent fashion. Among these associations (peaks) we focused our attention on those proximal to the regulatory region of HOTAIR and MALAT1. These transcripts appeared regulated by estrogens and able to control ERs function by interacting with ERα/ERβ as indicated by RNA-ChIP. Further studies performed by ChIRP revealed that in unstimulated condition, HOTAIR and MALAT1 were present on pS2, hTERT and HOTAIR promoters at the ERE/eNOS peaks. Interestingly, upon treatment with17β-estradiol HOTAIR recruitment to chromatin increased significantly while that of MALAT1 was reduced, suggesting an opposite regulation and function for these lncRNAs. Similar results were obtained in cells and in an ex vivo prostate organotypic slice cultures. Overall, our data provide evidence of a crosstalk between lncRNAs, estrogens and estrogen receptors in prostate cancer with important consequences on gene expression regulation.
Figures

References
-
- Tsai M. J. & O’Malley B. W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63, 451–486 (1994). - PubMed
-
- Carroll J. S. et al.. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

