Molecular mechanisms at the core of the plant circadian oscillator
- PMID: 27922614
- PMCID: PMC7750160
- DOI: 10.1038/nsmb.3327
Molecular mechanisms at the core of the plant circadian oscillator
Abstract
Circadian clocks are endogenous timekeeping networks that allow organisms to align their physiology with their changing environment and to perform biological processes at the most relevant times of the day and year. Initial feedback-loop models of the oscillator have been enriched by emerging evidence highlighting the increasing variety of factors and mechanisms that contribute to the generation of rhythms. In this Review, we consider the two major input pathways that connect the circadian clock of the model plant Arabidopsis thaliana to its environment and discuss recent advances in understanding of how transcriptional, post-translational and post-transcriptional mechanisms contribute to clock function.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
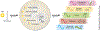

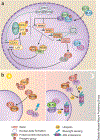
References
-
- Millar AJ The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol. 67, 595–618 (2016). This recent review provides a comprehensive discussion of plant circadian timekeeping circuits in the context of ecologically relevant environments. - PubMed
-
- Greenham K. & McClung CR Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet 16, 598–610 (2015). - PubMed
-
- Wang ZY & Tobin EM Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

