Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study
- PMID: 27923066
- PMCID: PMC5140047
- DOI: 10.1371/journal.pmed.1002162
Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study
Abstract
Background: Lung adenocarcinoma (LUAD) is the most common histologic subtype of lung cancer and has a high risk of distant metastasis at every disease stage. We aimed to characterize the genomic landscape of LUAD and identify mutation signatures associated with tumor progression.
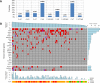
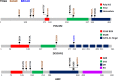
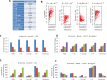
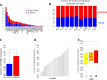
Methods and findings: We performed an integrative genomic analysis, incorporating whole exome sequencing (WES), determination of DNA copy number and DNA methylation, and transcriptome sequencing for 101 LUAD samples from the Environment And Genetics in Lung cancer Etiology (EAGLE) study. We detected driver genes by testing whether the nonsynonymous mutation rate was significantly higher than the background mutation rate and replicated our findings in public datasets with 724 samples. We performed subclonality analysis for mutations based on mutant allele data and copy number alteration data. We also tested the association between mutation signatures and clinical outcomes, including distant metastasis, survival, and tumor grade. We identified and replicated two novel candidate driver genes, POU class 4 homeobox 2 (POU4F2) (mutated in 9 [8.9%] samples) and ZKSCAN1 (mutated in 6 [5.9%] samples), and characterized their major deleterious mutations. ZKSCAN1 was part of a mutually exclusive gene set that included the RTK/RAS/RAF pathway genes BRAF, EGFR, KRAS, MET, and NF1, indicating an important driver role for this gene. Moreover, we observed strong associations between methylation in specific genomic regions and somatic mutation patterns. In the tumor evolution analysis, four driver genes had a significantly lower fraction of subclonal mutations (FSM), including TP53 (p = 0.007), KEAP1 (p = 0.012), STK11 (p = 0.0076), and EGFR (p = 0.0078), suggesting a tumor initiation role for these genes. Subclonal mutations were significantly enriched in APOBEC-related signatures (p < 2.5×10-50). The total number of somatic mutations (p = 0.0039) and the fraction of transitions (p = 5.5×10-4) were associated with increased risk of distant metastasis. Our study's limitations include a small number of LUAD patients for subgroup analyses and a single-sample design for investigation of subclonality.
Conclusions: These data provide a genomic characterization of LUAD pathogenesis and progression. The distinct clonal and subclonal mutation signatures suggest possible diverse carcinogenesis pathways for endogenous and exogenous exposures, and may serve as a foundation for more effective treatments for this lethal disease. LUAD's high heterogeneity emphasizes the need to further study this tumor type and to associate genomic findings with clinical outcomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

