Intact mitochondrial Ca2+ uniport is essential for agonist-induced activation of endothelial nitric oxide synthase (eNOS)
- PMID: 27923677
- PMCID: PMC5381715
- DOI: 10.1016/j.freeradbiomed.2016.11.049
Intact mitochondrial Ca2+ uniport is essential for agonist-induced activation of endothelial nitric oxide synthase (eNOS)
Abstract
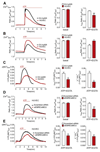
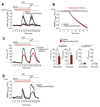
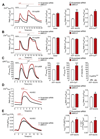
Mitochondrial Ca2+ uptake regulates diverse endothelial cell functions and has also been related to nitric oxide (NO•) production. However, it is not entirely clear if the organelles support or counteract NO• biosynthesis by taking up Ca2+. The objective of this study was to verify whether or not mitochondrial Ca2+ uptake influences Ca2+-triggered NO• generation by endothelial NO• synthase (eNOS) in an immortalized endothelial cell line (EA.hy926), respective primary human umbilical vein endothelial cells (HUVECs) and eNOS-RFP (red fluorescent protein) expressing human embryonic kidney (HEK293) cells. We used novel genetically encoded fluorescent NO• probes, the geNOps, and Ca2+ sensors to monitor single cell NO• and Ca2+ dynamics upon cell treatment with ATP, an inositol 1,4,5-trisphosphate (IP3)-generating agonist. Mitochondrial Ca2+ uptake was specifically manipulated by siRNA-mediated knock-down of recently identified key components of the mitochondrial Ca2+ uniporter machinery. In endothelial cells and the eNOS-RFP expressing HEK293 cells we show that reduced mitochondrial Ca2+ uptake upon the knock-down of the mitochondrial calcium uniporter (MCU) protein and the essential MCU regulator (EMRE) yield considerable attenuation of the Ca2+-triggered NO• increase independently of global cytosolic Ca2+ signals. The knock-down of mitochondrial calcium uptake 1 (MICU1), a gatekeeper of the MCU, increased both mitochondrial Ca2+ sequestration and Ca2+-induced NO• signals. The positive correlation between mitochondrial Ca2+ elevation and NO• production was independent of eNOS phosphorylation at serine1177. Our findings emphasize that manipulating mitochondrial Ca2+ uptake may represent a novel strategy to control eNOS-mediated NO• production.
Keywords: Calcium; ENOS; Endothelial nitric oxide production; GeNOps; Mitochondria.
Copyright © 2016. Published by Elsevier Inc.
Figures




Similar articles
-
Depolarization of mitochondria in endothelial cells promotes cerebral artery vasodilation by activation of nitric oxide synthase.Arterioscler Thromb Vasc Biol. 2013 Apr;33(4):752-9. doi: 10.1161/ATVBAHA.112.300560. Epub 2013 Jan 17. Arterioscler Thromb Vasc Biol. 2013. PMID: 23329133 Free PMC article.
-
Far-infrared radiation acutely increases nitric oxide production by increasing Ca(2+) mobilization and Ca(2+)/calmodulin-dependent protein kinase II-mediated phosphorylation of endothelial nitric oxide synthase at serine 1179.Biochem Biophys Res Commun. 2013 Jul 12;436(4):601-6. doi: 10.1016/j.bbrc.2013.06.003. Epub 2013 Jun 10. Biochem Biophys Res Commun. 2013. PMID: 23756809
-
Rearrangement of MICU1 multimers for activation of MCU is solely controlled by cytosolic Ca(2.).Sci Rep. 2015 Oct 22;5:15602. doi: 10.1038/srep15602. Sci Rep. 2015. PMID: 26489515 Free PMC article.
-
eNOS activation and NO function: pregnancy adaptive programming of capacitative entry responses alters nitric oxide (NO) output in vascular endothelium--new insights into eNOS regulation through adaptive cell signaling.J Endocrinol. 2011 Sep;210(3):243-58. doi: 10.1530/JOE-11-0053. Epub 2011 May 9. J Endocrinol. 2011. PMID: 21555345 Free PMC article. Review.
-
The mitochondrial Ca(2+) uniporter.Cell Calcium. 2012 Jul;52(1):16-21. doi: 10.1016/j.ceca.2012.04.006. Epub 2012 Jun 4. Cell Calcium. 2012. PMID: 22672876 Review.
Cited by
-
IKCa channels control breast cancer metabolism including AMPK-driven autophagy.Cell Death Dis. 2022 Oct 27;13(10):902. doi: 10.1038/s41419-022-05329-z. Cell Death Dis. 2022. PMID: 36302750 Free PMC article.
-
Modulatory Effect of Rosmarinic Acid on H2O2-Induced Adaptive Glycolytic Response in Dermal Fibroblasts.Molecules. 2023 Jul 24;28(14):5599. doi: 10.3390/molecules28145599. Molecules. 2023. PMID: 37513476 Free PMC article.
-
Sex Influence on Autophagy Markers and miRNAs in Basal and Angiotensin II-Treated Human Umbilical Vein Endothelial Cells.Int J Mol Sci. 2023 Oct 5;24(19):14929. doi: 10.3390/ijms241914929. Int J Mol Sci. 2023. PMID: 37834376 Free PMC article.
-
Endothelial MICU1 alleviates diabetic cardiomyopathy by attenuating nitrative stress-mediated cardiac microvascular injury.Cardiovasc Diabetol. 2023 Aug 17;22(1):216. doi: 10.1186/s12933-023-01941-1. Cardiovasc Diabetol. 2023. PMID: 37592255 Free PMC article.
-
Genetic biosensors for imaging nitric oxide in single cells.Free Radic Biol Med. 2018 Nov 20;128:50-58. doi: 10.1016/j.freeradbiomed.2018.01.027. Epub 2018 Feb 2. Free Radic Biol Med. 2018. PMID: 29398285 Free PMC article. Review.
References
-
- Denton RM. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta. 1787;2009:1309–1316. - PubMed
-
- Dedkova EN, Ji X, Lipsius SL, Blatter LA. Mitochondrial calcium uptake stimulates nitric oxide production in mitochondria of bovine vascular endothelial cells. Am J Physiol Cell Physiol. 2004;286:406–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

