RB Loss Promotes Prostate Cancer Metastasis
- PMID: 27923835
- PMCID: PMC5700768
- DOI: 10.1158/0008-5472.CAN-16-1589
RB Loss Promotes Prostate Cancer Metastasis
Abstract
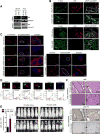
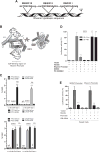
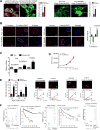
RB loss occurs commonly in neoplasia but its contributions to advanced cancer have not been assessed directly. Here we show that RB loss in multiple murine models of cancer produces a prometastatic phenotype. Gene expression analyses showed that regulation of the cell motility receptor RHAMM by the RB/E2F pathway was critical for epithelial-mesenchymal transition, motility, and invasion by cancer cells. Genetic modulation or pharmacologic inhibition of RHAMM activity was sufficient and necessary for metastatic phenotypes induced by RB loss in prostate cancer. Mechanistic studies in this setting established that RHAMM stabilized F-actin polymerization by controlling ROCK signaling. Collectively, our findings show how RB loss drives metastatic capacity and highlight RHAMM as a candidate therapeutic target for treating advanced prostate cancer. Cancer Res; 77(4); 982-95. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

