Accounting for Protein Subcellular Localization: A Compartmental Map of the Rat Liver Proteome
- PMID: 27923875
- PMCID: PMC5294208
- DOI: 10.1074/mcp.M116.064527
Accounting for Protein Subcellular Localization: A Compartmental Map of the Rat Liver Proteome
Abstract
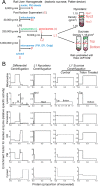
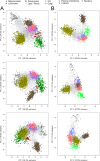
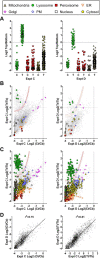
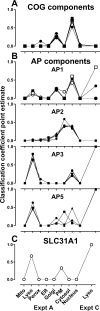
Accurate knowledge of the intracellular location of proteins is important for numerous areas of biomedical research including assessing fidelity of putative protein-protein interactions, modeling cellular processes at a system-wide level and investigating metabolic and disease pathways. Many proteins have not been localized, or have been incompletely localized, partly because most studies do not account for entire subcellular distribution. Thus, proteins are frequently assigned to one organelle whereas a significant fraction may reside elsewhere. As a step toward a comprehensive cellular map, we used subcellular fractionation with classic balance sheet analysis and isobaric labeling/quantitative mass spectrometry to assign locations to >6000 rat liver proteins. We provide quantitative data and error estimates describing the distribution of each protein among the eight major cellular compartments: nucleus, mitochondria, lysosomes, peroxisomes, endoplasmic reticulum, Golgi, plasma membrane and cytosol. Accounting for total intracellular distribution improves quality of organelle assignments and assigns proteins with multiple locations. Protein assignments and supporting data are available online through the Prolocate website (http://prolocate.cabm.rutgers.edu). As an example of the utility of this data set, we have used organelle assignments to help analyze whole exome sequencing data from an infant dying at 6 months of age from a suspected neurodegenerative lysosomal storage disorder of unknown etiology. Sequencing data was prioritized using lists of lysosomal proteins comprising well-established residents of this organelle as well as novel candidates identified in this study. The latter included copper transporter 1, encoded by SLC31A1, which we localized to both the plasma membrane and lysosome. The patient harbors two predicted loss of function mutations in SLC31A1, suggesting that this may represent a heretofore undescribed recessive lysosomal storage disease gene.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Rolland T., Tasan M., Charloteaux B., Pevzner S. J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., Kamburov A., Ghiassian S. D., Yang X., Ghamsari L., Balcha D., Begg B. E., Braun P., Brehme M., Broly M. P., Carvunis A. R., Convery-Zupan D., Corominas R., Coulombe-Huntington J., Dann E., Dreze M., Dricot A., Fan C., Franzosa E., Gebreab F., Gutierrez B. J., Hardy M. F., Jin M., Kang S., Kiros R., Lin G. N., Luck K., MacWilliams A., Menche J., Murray R. R., Palagi A., Poulin M. M., Rambout X., Rasla J., Reichert P., Romero V., Ruyssinck E., Sahalie J. M., Scholz A., Shah A. A., Sharma A., Shen Y., Spirohn K., Tam S., Tejeda A. O., Trigg S. A., Twizere J. C., Vega K., Walsh J., Cusick M. E., Xia Y., Barabasi A. L., Iakoucheva L. M., Aloy P., De Las Rivas J., Tavernier J., Calderwood M. A., Hill D. E., Hao T., Roth F. P., and Vidal M. (2014) A proteome-scale map of the human interactome network. Cell 159, 1212–1226 - PMC - PubMed
-
- Bradbury A., and Pluckthun A. (2015) Reproducibility: Standardize antibodies used in research. Nature 518, 27–29 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

