Antiviral Innate Immune Response Interferes with the Formation of Replication-Associated Membrane Structures Induced by a Positive-Strand RNA Virus
- PMID: 27923923
- PMCID: PMC5142621
- DOI: 10.1128/mBio.01991-16
Antiviral Innate Immune Response Interferes with the Formation of Replication-Associated Membrane Structures Induced by a Positive-Strand RNA Virus
Abstract
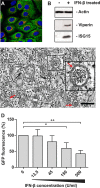
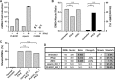
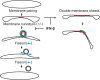
Infection with nidoviruses like corona- and arteriviruses induces a reticulovesicular network of interconnected endoplasmic reticulum (ER)-derived double-membrane vesicles (DMVs) and other membrane structures. This network is thought to accommodate the viral replication machinery and protect it from innate immune detection. We hypothesized that the innate immune response has tools to counteract the formation of these virus-induced replication organelles in order to inhibit virus replication. Here we have investigated the effect of type I interferon (IFN) treatment on the formation of arterivirus-induced membrane structures. Our approach involved ectopic expression of arterivirus nonstructural proteins nsp2 and nsp3, which induce DMV formation in the absence of other viral triggers of the interferon response, such as replicating viral RNA. Thus, this setup can be used to identify immune effectors that specifically target the (formation of) virus-induced membrane structures. Using large-scale electron microscopy mosaic maps, we found that IFN-β treatment significantly reduced the formation of the membrane structures. Strikingly, we also observed abundant stretches of double-membrane sheets (a proposed intermediate of DMV formation) in IFN-β-treated samples, suggesting the disruption of DMV biogenesis. Three interferon-stimulated gene products, two of which have been reported to target the hepatitis C virus replication structures, were tested for their possible involvement, but none of them affected membrane structure formation. Our study reveals the existence of a previously unknown innate immune mechanism that antagonizes the viral hijacking of host membranes. It also provides a solid basis for further research into the poorly understood interactions between the innate immune system and virus-induced replication structures.
Importance: Viruses with a positive-strand RNA genome establish a membrane-associated replication organelle by hijacking and remodeling intracellular host membranes, a process deemed essential for their efficient replication. It is unknown whether the cellular innate immune system can detect and/or inhibit the formation of these membrane structures, which could be an effective mechanism to delay viral RNA replication. In this study, using an expression system that closely mimics the formation of arterivirus replication structures, we show for the first time that IFN-β treatment clearly reduces the amount of induced membrane structures. Moreover, drastic morphological changes were observed among the remaining structures, suggesting that their biogenesis was impaired. Follow-up experiments suggested that host cells contain a hitherto unknown innate antiviral mechanism, which targets this common feature of positive-strand RNA virus replication. Our study provides a strong basis for further research into the interaction of the innate immune system with membranous viral replication organelles.
Copyright © 2016 Oudshoorn et al.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
