FARNA: knowledgebase of inferred functions of non-coding RNA transcripts
- PMID: 27924038
- PMCID: PMC5389649
- DOI: 10.1093/nar/gkw973
FARNA: knowledgebase of inferred functions of non-coding RNA transcripts
Abstract
Non-coding RNA (ncRNA) genes play a major role in control of heterogeneous cellular behavior. Yet, their functions are largely uncharacterized. Current available databases lack in-depth information of ncRNA functions across spectrum of various cells/tissues. Here, we present FARNA, a knowledgebase of inferred functions of 10,289 human ncRNA transcripts (2,734 microRNA and 7,555 long ncRNA) in 119 tissues and 177 primary cells of human. Since transcription factors (TFs) and TF co-factors (TcoFs) are crucial components of regulatory machinery for activation of gene transcription, cellular processes and diseases in which TFs and TcoFs are involved suggest functions of the transcripts they regulate. In FARNA, functions of a transcript are inferred from TFs and TcoFs whose genes co-express with the transcript controlled by these TFs and TcoFs in a considered cell/tissue. Transcripts were annotated using statistically enriched GO terms, pathways and diseases across cells/tissues based on guilt-by-association principle. Expression profiles across cells/tissues based on Cap Analysis of Gene Expression (CAGE) are provided. FARNA, having the most comprehensive function annotation of considered ncRNAs across widest spectrum of human cells/tissues, has a potential to greatly contribute to our understanding of ncRNA roles and their regulatory mechanisms in human. FARNA can be accessed at: http://cbrc.kaust.edu.sa/farna.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
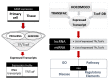

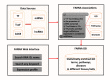
References
-
- Mattick J.S., Makunin I.V.. Non-coding RNA. Hum. Mol. Genet. 2006; 15:R17–R29. - PubMed
-
- Mercer T.R., Dinger M.E., Mattick J.S.. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009; 10:155–159. - PubMed
-
- Long J., Ou C., Xia H., Zhu Y., Liu D.. MiR-503 inhibited cell proliferation of human breast cancer cells by suppressing CCND1 expression. Tumour Biol. 2015; 36:8697–8702. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

