Metabolic regulation of gene expression through histone acylations
- PMID: 27924077
- PMCID: PMC5320945
- DOI: 10.1038/nrm.2016.140
Metabolic regulation of gene expression through histone acylations
Abstract
Eight types of short-chain Lys acylations have recently been identified on histones: propionylation, butyrylation, 2-hydroxyisobutyrylation, succinylation, malonylation, glutarylation, crotonylation and β-hydroxybutyrylation. Emerging evidence suggests that these histone modifications affect gene expression and are structurally and functionally different from the widely studied histone Lys acetylation. In this Review, we discuss the regulation of non-acetyl histone acylation by enzymatic and metabolic mechanisms, the acylation 'reader' proteins that mediate the effects of different acylations and their physiological functions, which include signal-dependent gene activation, spermatogenesis, tissue injury and metabolic stress. We propose a model to explain our present understanding of how differential histone acylation is regulated by the metabolism of the different acyl-CoA forms, which in turn modulates the regulation of gene expression.
Figures



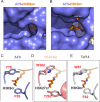
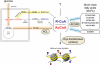
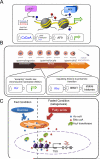
Comment in
-
HATs off for the Lasker awardees.Nat Rev Mol Cell Biol. 2018 Nov;19(11):677. doi: 10.1038/s41580-018-0065-3. Nat Rev Mol Cell Biol. 2018. PMID: 30232395 No abstract available.
References
-
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

