Smoking-associated DNA methylation markers predict lung cancer incidence
- PMID: 27924164
- PMCID: PMC5123284
- DOI: 10.1186/s13148-016-0292-4
Smoking-associated DNA methylation markers predict lung cancer incidence
Abstract
Background: Newly established blood DNA methylation markers that are strongly associated with smoking might open new avenues for lung cancer (LC) screening. We aimed to assess the performance of the top hits from previous epigenome-wide association studies in prediction of LC incidence. In a prospective nested case-control study, DNA methylation at AHRR (cg05575921), 6p21.33 (cg06126421), and F2RL3 (cg03636183) were measured by pyrosequencing in baseline whole blood samples of 143 incident LC cases identified during 11 years of follow-up and 457 age- and sex-matched controls without diagnosis of LC until the end of follow-up. The individual and joint associations of the 3 markers with LC risk were estimated by logistic regression, adjusted for potential confounders including smoking status and cigarette pack-years. The predictive performance was evaluated for both the individual markers and their combinations derived from multiple algorithms.
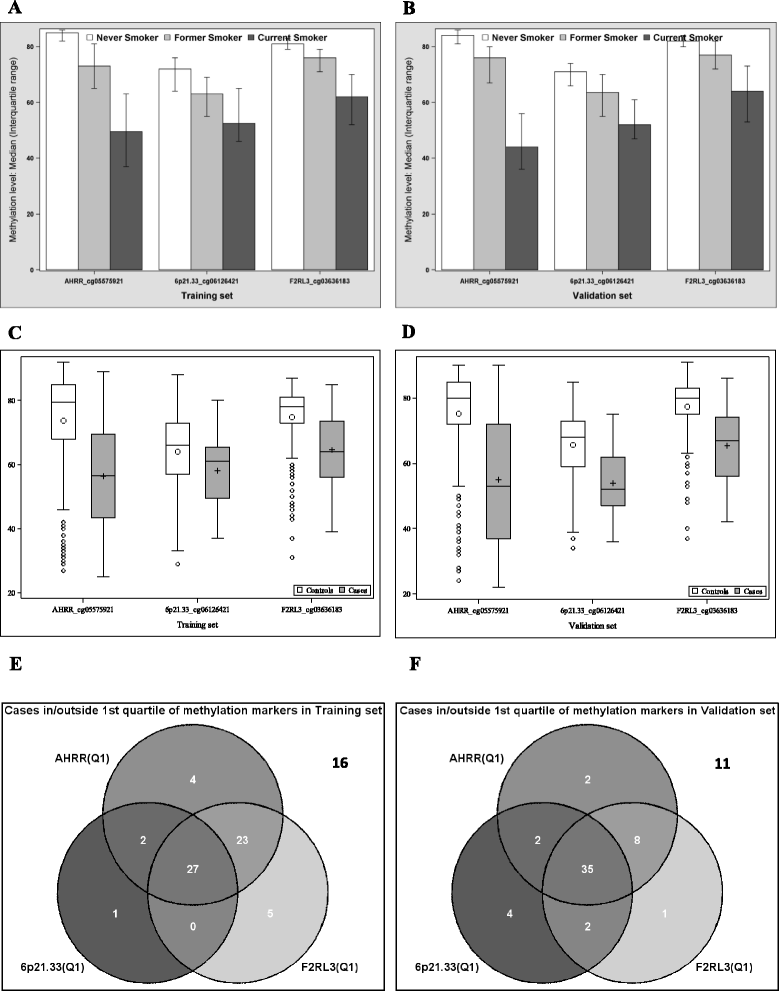
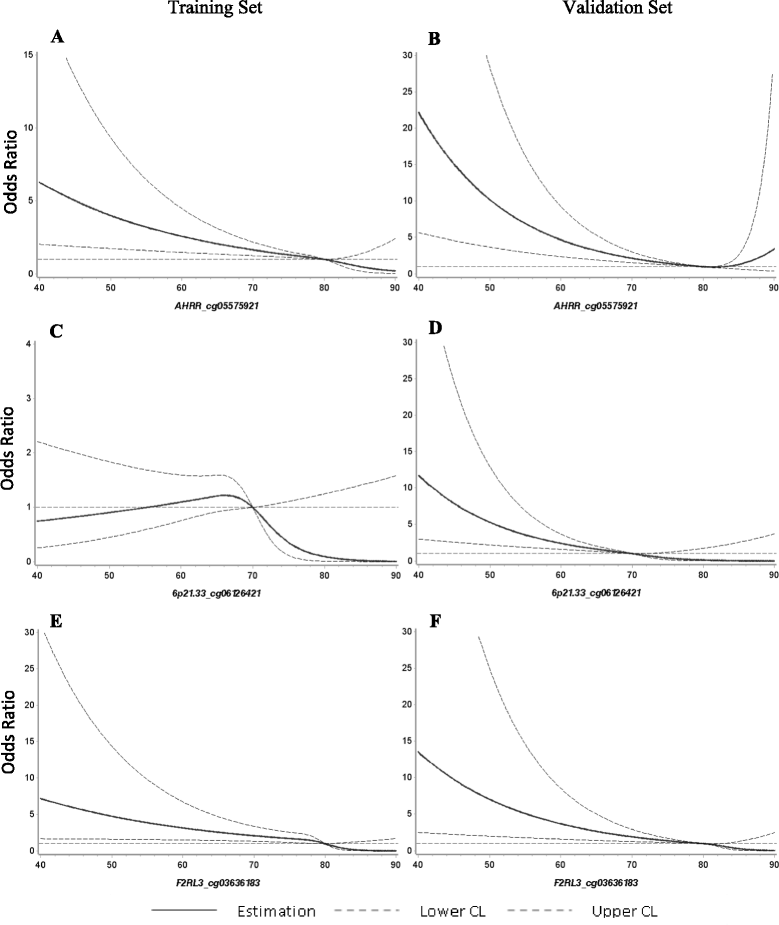
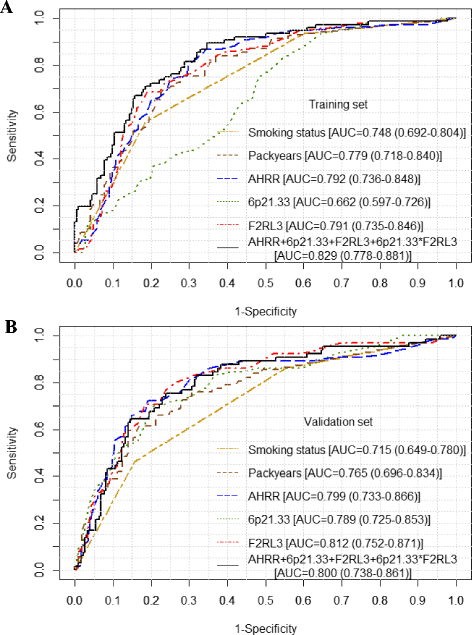
Results: Pronounced demethylation of all 3 markers was observed at baseline among cases compared to controls. Risk of developing LC increased with decreasing DNA methylation levels, with adjusted ORs (95% CI) of 15.86 (4.18-60.17), 8.12 (2.69-4.48), and 10.55 (3.44-32.31), respectively, for participants in the lowest quartile of AHRR, 6p21.33, and F2RL3 compared to participants in the highest 2 quartiles of each site among controls. The individual 3 markers exhibited similar accuracy in predicting LC incidence, with AUCs ranging from 0.79 to 0.81. Combination of the 3 markers did not improve the predictive performance (AUC = 0.80). The individual markers or their combination outperformed self-reported smoking exposure particularly in light smokers. No variation in risk prediction was identified with respect to age, follow-up time, and histological subtypes.
Conclusions: AHRR, 6p21.33, and F2RL3 methylation in blood DNA are predictive for LC development, which might be useful for identification of risk groups for further specific screening, such as CT examination.
Keywords: AHRR; DNA methylation; F2RL3; Lung cancer; Risk prediction; Smoking.
Figures

References
-
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase no. 11. Lyon, France: International Agency for Research on Cancer; 2013.
-
- Howlader N, Noone A, Krapcho M, Garshell J, Miller D, Altekruse S, et al. SEER cancer statistics review, 1975-2012, National Cancer Institute. 2015. Available from: http://seer.cancer.gov/csr/1975_2012/. Accessed Jan 2016.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

