Insights into the evolution of pathogenicity of Escherichia coli from genomic analysis of intestinal E. coli of Marmota himalayana in Qinghai-Tibet plateau of China
- PMID: 27924811
- PMCID: PMC5180367
- DOI: 10.1038/emi.2016.122
Insights into the evolution of pathogenicity of Escherichia coli from genomic analysis of intestinal E. coli of Marmota himalayana in Qinghai-Tibet plateau of China
Abstract
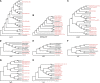
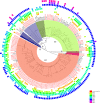
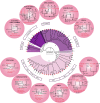
Escherichia coli is both of a widespread harmless gut commensal and a versatile pathogen of humans. Domestic animals are a well-known reservoir for pathogenic E. coli. However, studies of E. coli populations from wild animals that have been separated from human activities had been very limited. Here we obtained 580 isolates from intestinal contents of 116 wild Marmot Marmota himalayana from Qinghai-Tibet plateau, China, with five isolates per animal. We selected 125 (hereinafter referred to as strains) from the 580 isolates for genome sequencing, based on unique pulse field gel electrophoresis patterns and at least one isolate per animal. Whole genome sequence analysis revealed that all 125 strains carried at least one and the majority (79.2%) carried multiple virulence genes based on the analysis of 22 selected virulence genes. In particular, the majority of the strains carried virulence genes from different pathovars as potential 'hybrid pathogens'. The alleles of eight virulence genes from the Marmot E. coli were found to have diverged earlier than all known alleles from human and other animal E. coli. Phylogenetic analysis of the 125 Marmot E. coli genomes and 355 genomes selected from 1622 human and other E. coli strains identified two new phylogroups, G and H, both of which diverged earlier than the other phylogroups. Eight of the 12 well-known pathogenic E. coli lineages were found to share a most recent common ancestor with one or more Marmot E. coli strains. Our results suggested that the intestinal E. coli of the Marmots contained a diverse virulence gene pool and is potentially pathogenic to humans. These findings provided a new understanding of the evolutionary origin of pathogenic E. coli.
Figures

Similar articles
-
Genomic and molecular characterisation of Escherichia marmotae from wild rodents in Qinghai-Tibet plateau as a potential pathogen.Sci Rep. 2019 Jul 23;9(1):10619. doi: 10.1038/s41598-019-46831-3. Sci Rep. 2019. PMID: 31337784 Free PMC article.
-
Comparative genomic analysis of 127 Escherichia coli strains isolated from domestic animals with diarrhea in China.BMC Genomics. 2019 Mar 13;20(1):212. doi: 10.1186/s12864-019-5588-2. BMC Genomics. 2019. PMID: 30866824 Free PMC article.
-
Genomic Epidemiology and Evolution of Escherichia coli in Wild Animals in Mexico.mSphere. 2021 Jan 6;6(1):e00738-20. doi: 10.1128/mSphere.00738-20. mSphere. 2021. PMID: 33408222 Free PMC article.
-
Virulence potential for extraintestinal infections among commensal Escherichia coli isolated from healthy humans--the Trojan horse within our gut.FEMS Microbiol Lett. 2015 Mar;362(5):fnu061. doi: 10.1093/femsle/fnu061. Epub 2015 Feb 5. FEMS Microbiol Lett. 2015. PMID: 25657191 Review.
-
What defines extraintestinal pathogenic Escherichia coli?Int J Med Microbiol. 2011 Dec;301(8):642-7. doi: 10.1016/j.ijmm.2011.09.006. Epub 2011 Oct 5. Int J Med Microbiol. 2011. PMID: 21982038 Review.
Cited by
-
ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping.Microb Genom. 2018 Jul;4(7):e000192. doi: 10.1099/mgen.0.000192. Epub 2018 Jun 19. Microb Genom. 2018. PMID: 29916797 Free PMC article.
-
Genomic and molecular characterisation of Escherichia marmotae from wild rodents in Qinghai-Tibet plateau as a potential pathogen.Sci Rep. 2019 Jul 23;9(1):10619. doi: 10.1038/s41598-019-46831-3. Sci Rep. 2019. PMID: 31337784 Free PMC article.
-
Stratified reconstruction of ancestral Escherichia coli diversification.BMC Genomics. 2019 Dec 5;20(1):936. doi: 10.1186/s12864-019-6346-1. BMC Genomics. 2019. PMID: 31805853 Free PMC article.
-
Escherichia marmotae-a Human Pathogen Easily Misidentified as Escherichia coli.Microbiol Spectr. 2022 Apr 27;10(2):e0203521. doi: 10.1128/spectrum.02035-21. Epub 2022 Apr 5. Microbiol Spectr. 2022. PMID: 35380461 Free PMC article.
-
Decomposition of the pangenome matrix reveals a structure in gene distribution in the Escherichia coli species.mSphere. 2025 Jan 28;10(1):e0053224. doi: 10.1128/msphere.00532-24. Epub 2024 Dec 31. mSphere. 2025. PMID: 39745367 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
