Posttranslational Modifications of Lipid-Activated Nuclear Receptors: Focus on Metabolism
- PMID: 27925773
- PMCID: PMC5413085
- DOI: 10.1210/en.2016-1577
Posttranslational Modifications of Lipid-Activated Nuclear Receptors: Focus on Metabolism
Abstract
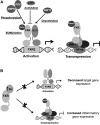
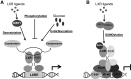
Posttranslational modifications (PTMs) occur to nearly all proteins, are catalyzed by specific enzymes, and are subjected to tight regulation. They have been shown to be a powerful means by which the function of proteins can be modified, resulting in diverse effects. Technological advances such as the increased sensitivity of mass spectrometry-based techniques and availability of mutant animal models have enhanced our understanding of the complexities of their regulation and the effect they have on protein function. However, the role that PTMs have in a pathological context still remains unknown for the most part. PTMs enable the modulation of nuclear receptor function in a rapid and reversible manner in response to varied stimuli, thereby dramatically altering their activity in some cases. This review focuses on acetylation, phosphorylation, SUMOylation, and O-GlcNAcylation, which are the 4 most studied PTMs affecting lipid-regulated nuclear receptor biology, as well as on the implications of such modifications on metabolic pathways under homeostatic and pathological situations. Moreover, we review recent studies on the modulation of PTMs as therapeutic targets for metabolic diseases.
Figures

References
-
- Lu C-T, Huang K-Y, Su M-G, Lee T-Y, Bretaña NA, Chang W-C, Chen Y-J, Chen Y-J, Huang H-D. DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res. 2013;41(database issue):D295–D305. - PMC - PubMed
-
- Ristic M, Brockly F, Piechaczyk M, Bossis G. Detection of protein–protein interactions and posttranslational modifications using the proximity ligation assay: application to the study of the SUMO pathway. Methods Mol Biol. 2016;1449:279–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

